Circular RNA-mediated miRNA sponge & RNA binding protein in biological modulation of breast cancer
- PMID: 38282696
- PMCID: PMC10818160
- DOI: 10.1016/j.ncrna.2023.12.005
Circular RNA-mediated miRNA sponge & RNA binding protein in biological modulation of breast cancer
Abstract
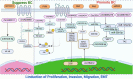
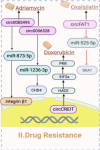
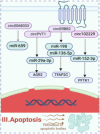
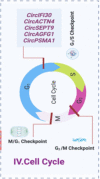
Circular RNAs (circRNAs) and small non-coding RNAs of the head-to-junction circle in the construct play critical roles in gene regulation and are significantly associated with breast cancer (BC). Numerous circRNAs are potential cancer biomarkers that may be used for diagnosis and prognosis. Widespread expression of circRNAs is regarded as a feature of gene expression in highly diverged eukaryotes. Recent studies show that circRNAs have two main biological modulation models: sponging and RNA-binding. This review explained the biogenesis of circRNAs and assessed emerging findings on their sponge function and role as RNA-binding proteins (RBPs) to better understand how their interaction alters cellular function in BC. We focused on how sponges significantly affect the phenotype and progression of BC. We described how circRNAs exercise the translation functions in ribosomes. Furthermore, we reviewed recent studies on RBPs, and post-protein modifications influencing BC and provided a perspective on future research directions for treating BC.
Keywords: Biogenesis; Breast cancer; CircRNAs; RNA-Binding proteins (RBPs); Sponge.
© 2024 The Authors.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

References
Publication types
LinkOut - more resources
Full Text Sources

