The complete chloroplast genome of Meconopsis bella Prain 1894 (Papaveraceae), a high-altitude plant distributed on the Qinghai-Tibet plateau
- PMID: 38282978
- PMCID: PMC10812850
- DOI: 10.1080/23802359.2024.2306879
The complete chloroplast genome of Meconopsis bella Prain 1894 (Papaveraceae), a high-altitude plant distributed on the Qinghai-Tibet plateau
Abstract
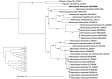
Meconopsis bella Prain 1894 (M. bella) is a rare herb within the family Papaveraceae of which unique and gorgeous purple flowers are blooming in the flowering phase. In this study, we reported the complete chloroplast (cp) genome of M. bella, which was mainly distributed on the Qinghai-Tibet plateau. The complete chloroplast genome sequence of M. bella was 153,073 bp in size and was characterized by a typical quadripartite structure consisting of a large single-copy (LSC) region of 83,562 bp, a small single-copy (SSC) region of 178,33 bp and two identical inverted repeats (IR) regions of 25,839 bp. The genome contained 133 genes, including 88 protein-encoding genes, eight ribosomal RNA genes, and 37 transfer RNA genes. Phylogenetic analysis based on the maximum-likelihood (ML) method showed that M. bella was closely related to M. paniculate and M. pinnatifolia within the genus Meconopsis.
Keywords: Meconopsis bella; Qinghai-Tibet plateau; chloroplast genome; papaveraceae.
© 2024 The Author(s). Published by Informa UK Limited, trading as Taylor & Francis Group.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures


References
-
- Jun-Hua DU, Wen-Jing Z, Ji-Rong LI, Ying LI.. 2011. The overview of exploiture of the curatorial and ornamental resources on Wild Flora Meconopsis Vig. J Qinghai Norm Univ(Natural Science Ed). 27(04):52–57.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
