In Silico Identification and Characterization of Drug Targets in Streptococcus pneumoniae ATCC 700669 (Serotype 23F) by Subtractive Genomics
- PMID: 38283072
- PMCID: PMC10821801
- DOI: 10.1155/2024/5917667
In Silico Identification and Characterization of Drug Targets in Streptococcus pneumoniae ATCC 700669 (Serotype 23F) by Subtractive Genomics
Abstract
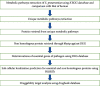
Streptococcus pneumoniae (S. pneumoniae) is an important pathogen worldwide that causes pneumococcal infections which are related to high rates of morbidity and mortality especially in young children, older adults, and immune-compromised persons. Antibiotic resistance in S. pneumoniae is a serious problem across the world from time to time, resulting in treatment failure and diminished value of older medicines. Therefore, the objective of this study was to identify new putative drug targets against S. pneumoniae serotype 23F by using subtractive genomics. By using bioinformatics tools such as NCBI, UniProt KB, PDB, KEGG, DEG, PSORTb, CD hit, DrugBank database, and other softwares, proteins involved in unique metabolic pathways of S. pneumoniae serotype 23F were studied. The result indicates that this serotype consists of 97 metabolic pathways of which 74 are common with that of human, and 23 pathways are unique to the serotype 23F. After investigation and analysis of essentiality, nonhomology, subcellular localization, having drug targets, and enzymatic activity, four proteins were prioritized as druggable targets. These druggable proteins include UDP-N-acetylglucosamine 1-carboxyvinyltransferase, UDP-N-acetyl muramate dehydrogenase, D-alanine-D-alanine ligase, and alanine racemase that are found in S. pneumoniae serotype 23F. All these four proteins are essential, are nonhomologous with human proteins, have drug targets, and are located in cell cytoplasm. Therefore, the authors recommend these proteins to be used for efficient drug design against S. pneumoniae serotype 23F after experimental validation for essentiality and druggability.
Copyright © 2024 Tolossa Duguma and Hunduma Dinka.
Conflict of interest statement
The authors declare that there are no conflicts of interest regarding the publication of this paper.
Figures
References
-
- World Health Organization. Antimicrobial Resistance: Global Report on Surveillance: Antimicrobial Resistance Global surveillance Report . Geneva: World Health Organization; 2014.
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases