Systems biology approach to identify biomarkers and therapeutic targets for colorectal cancer
- PMID: 38283191
- PMCID: PMC10821538
- DOI: 10.1016/j.bbrep.2023.101633
Systems biology approach to identify biomarkers and therapeutic targets for colorectal cancer
Abstract
Background: Colorectal cancer (CRC), is the third most prevalent cancer across the globe, and is often detected at advanced stage. Late diagnosis of CRC, leave the chemotherapy and radiotherapy as the main options for the possible treatment of the disease which are associated with severe side effects. In the present study, we seek to explore CRC gene expression data using a systems biology framework to identify potential biomarkers and therapeutic targets for earlier diagnosis and treatment of the disease.
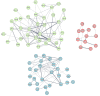
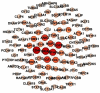
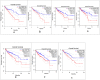
Methods: The expression data was retrieved from the gene expression omnibus (GEO). Differential gene expression analysis was conducted using R/Bioconductor package. The PPI network was reconstructed by the STRING. Cystoscope and Gephi software packages were used for visualization and centrality analysis of the PPI network. Clustering analysis of the PPI network was carried out using k-mean algorithm. Gene-set enrichment based on Gene Ontology (GO) and KEGG pathway databases was carried out to identify the biological functions and pathways associated with gene groups. Prognostic value of the selected identified hub genes was examined by survival analysis, using GEPIA.
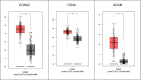
Results: A total of 848 differentially expressed genes were identified. Centrality analysis of the PPI network resulted in identification of 99 hubs genes. Clustering analysis dissected the PPI network into seven interactive modules. While several DEGs and the central genes in each module have already reported to contribute to CRC progression, survival analysis confirmed high expression of central genes, CCNA2, CD44, and ACAN contribute to poor prognosis of CRC patients. In addition, high expression of TUBA8, AMPD3, TRPC1, ARHGAP6, JPH3, DYRK1A and ACTA1 was found to associate with decreased survival rate.
Conclusion: Our results identified several genes with high centrality in PPI network that contribute to progression of CRC. The fact that several of the identified genes have already been reported to be relevant to diagnosis and treatment of CRC, other highlighted genes with limited literature information may hold potential to be explored in the context of CRC biomarker and drug target discovery.
Keywords: Biomarker; Colorectal cancer; Hub gene; PPI network.
© 2023 The Authors.
Conflict of interest statement
None.
Figures

Similar articles
-
Identification of potential biomarkers with colorectal cancer based on bioinformatics analysis and machine learning.Math Biosci Eng. 2021 Oct 19;18(6):8997-9015. doi: 10.3934/mbe.2021443. Math Biosci Eng. 2021. PMID: 34814332
-
Identification of key genes for predicting colorectal cancer prognosis by integrated bioinformatics analysis.Oncol Lett. 2020 Jan;19(1):388-398. doi: 10.3892/ol.2019.11068. Epub 2019 Nov 7. Oncol Lett. 2020. PMID: 31897151 Free PMC article.
-
Bioinformatic Identification of Hub Genes and Analysis of Prognostic Values in Colorectal Cancer.Nutr Cancer. 2021;73(11-12):2568-2578. doi: 10.1080/01635581.2020.1841249. Epub 2020 Nov 5. Nutr Cancer. 2021. PMID: 33153324
-
Bioinformatics-based identification of key genes and pathways associated with colorectal cancer diagnosis, treatment, and prognosis.Medicine (Baltimore). 2022 Sep 16;101(37):e30619. doi: 10.1097/MD.0000000000030619. Medicine (Baltimore). 2022. PMID: 36123948 Free PMC article.
-
Common gene signatures and key pathways in hypopharyngeal and esophageal squamous cell carcinoma: Evidence from bioinformatic analysis.Medicine (Baltimore). 2020 Oct 16;99(42):e22434. doi: 10.1097/MD.0000000000022434. Medicine (Baltimore). 2020. PMID: 33080677 Free PMC article.
Cited by
-
Pan-cancer analysis reveals immunological and prognostic significance of CCT5 in human tumors.Sci Rep. 2025 Apr 24;15(1):14405. doi: 10.1038/s41598-025-88339-z. Sci Rep. 2025. PMID: 40274875 Free PMC article.
References
-
- Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2018;68(6):394–424. - PubMed
-
- Favoriti P., Carbone G., Greco M., Pirozzi F., Pirozzi R.E.M., Corcione F. Worldwide burden of colorectal cancer: a review. Updates Surg. 2016;68:7–11. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

