Genome size variation and evolution during invasive range expansion in an introduced plant
- PMID: 38283607
- PMCID: PMC10810172
- DOI: 10.1111/eva.13624
Genome size variation and evolution during invasive range expansion in an introduced plant
Abstract
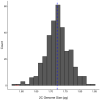
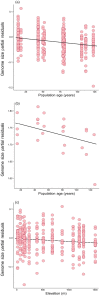
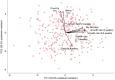
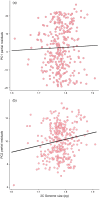
Plants demonstrate exceptional variation in genome size across species, and their genome sizes can also vary dramatically across individuals and populations within species. This aspect of genetic variation can have consequences for traits and fitness, but few studies attributed genome size differentiation to ecological and evolutionary processes. Biological invasions present particularly useful natural laboratories to infer selective agents that might drive genome size shifts across environments and population histories. Here, we test hypotheses for the evolutionary causes of genome size variation across 14 invading populations of yellow starthistle, Centaurea solstitialis, in California, United States. We use a survey of genome sizes and trait variation to ask: (1) Is variation in genome size associated with developmental trait variation? (2) Are genome sizes smaller toward the leading edge of the expansion, consistent with selection for "colonizer" traits? Or alternatively, does genome size increase toward the leading edge of the expansion, consistent with predicted consequences of founder effects and drift? (3) Finally, are genome sizes smaller at higher elevations, consistent with selection for shorter development times? We found that 2C DNA content varied 1.21-fold among all samples, and was associated with flowering time variation, such that plants with larger genomes reproduced later, with lower lifetime capitula production. Genome sizes increased toward the leading edge of the invasion, but tended to decrease at higher elevations, consistent with genetic drift during range expansion but potentially strong selection for smaller genomes and faster development time at higher elevations. These results demonstrate how genome size variation can contribute to traits directly tied to reproductive success, and how selection and drift can shape that variation. We highlight the influence of genome size on dynamics underlying a rapid range expansion in a highly problematic invasive plant.
Keywords: flow cytometry; intraspecific genome size variation; invasive species; yellow starthistle.
© 2023 The Authors. Evolutionary Applications published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Achigan‐Dako, E. G. , Fuchs, J. , Ahanchede, A. , & Blattner, F. R. (2008). Flow cytometric analysis in Lagenaria siceraria (Cucurbitaceae) indicates correlation of genome size with usage types and growing elevation. Plant Systematics and Evolution, 276(1–2), 9–19. 10.1007/s00606-008-0075-2 - DOI
-
- Baker, H. G. (1974). The evolution of weeds. Annual Review of Ecology and Systematics, 5(1), 1–24.
-
- Bancheva, S. , & Greilhuber, J. (2006). Genome size in Bulgarian Centaurea s.l. (Asteraceae). Plant Systematics and Evolution, 257(1–2), 95–117. 10.1007/s00606-005-0384-7 - DOI
-
- Barker, B. S. , Andonian, K. , Swope, S. M. , Luster, D. G. , & Dlugosch, K. M. (2017). Population genomic analyses reveal a history of range expansion and trait evolution across the native and invaded range of yellow starthistle (Centaurea solstitialis). Molecular Ecology, 26(4), 1131–1147. 10.1111/mec.13998 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

