Impact of platelet-derived mitochondria transfer in the metabolic profiling and progression of metastatic MDA-MB-231 human triple-negative breast cancer cells
- PMID: 38283990
- PMCID: PMC10811077
- DOI: 10.3389/fcell.2023.1324158
Impact of platelet-derived mitochondria transfer in the metabolic profiling and progression of metastatic MDA-MB-231 human triple-negative breast cancer cells
Abstract
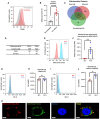
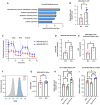
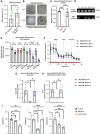
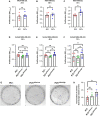
Introduction: An active role of platelets in the progression of triple-negative breast cancer (TNBC) cells has been described. Even the role of platelet-derived extracellular vesicles on the migration of MDA-MB-231 cells has been reported. Interestingly, upon activation, platelets release functional mitochondria into the extracellular environment. However, the impact of these platelet-derived mitochondria on the metabolic properties of MDA-MB-231 cells remains unclear. Methods: MDA-MB-231 and MDA-MB-231-Rho-0 cells were co-cultured with platelets, which were isolated from donor blood. Mitochondrial transfer was assessed through confocal microscopy and flow cytometry, while metabolic analyses were conducted using a Seahorse XF HS Mini Analyzer. The mito-chondrial DNA (mtDNA) copy number was determined via quantitative PCR (qPCR) following platelet co-culture. Finally, cell proliferation and colony formation assay were performed using crystal violet staining. Results and Discussion: We have shown that platelet-derived mitochondria are internalized by MDA-MB-231 cells in co-culture with platelets, increasing ATP production, oxygen (O2) consumption rate (OCR), cell proliferation, and metabolic adaptability. Additionally, we observed that MDA-MB-231 cells depleted from mtDNA restore cell proliferation in uridine/pyruvate-free cell culture medium and mitochondrial O2 consumption after co-culture with platelets, indicating a reconstitution of mtDNA facilitated by platelet-derived mitochondria. In conclusion, our study provides new insights into the role of platelet-derived mitochondria in the metabolic adaptability and progression of metastatic MDA-MB-231 TNBC cells.
Keywords: cancer; metabolic adaptability; mitochondria transfer; platelet-derived mitochondria; platelets.
Copyright © 2024 Cereceda, Cardenas, Khoury, Silva-Pavez and Hidalgo.
Conflict of interest statement
MK is the chief scientific officer of Cells for Cells and Regenero, the Chilean Consortium for Regenerative Medicine. The remaining authors declare that the research was conducted without commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures
Similar articles
-
Overexpression of multiple epidermal growth factor like domains 11 rescues anoikis survival through tumor cells-platelet interaction in triple negative breast Cancer cells.Life Sci. 2022 Jun 15;299:120541. doi: 10.1016/j.lfs.2022.120541. Epub 2022 Apr 7. Life Sci. 2022. PMID: 35398017
-
Combination of sirtuin 3 and hyperoxia diminishes tumorigenic properties of MDA-MB-231 cells.Life Sci. 2020 Aug 1;254:117812. doi: 10.1016/j.lfs.2020.117812. Epub 2020 May 16. Life Sci. 2020. PMID: 32428596
-
Metabolic Response of Triple-Negative Breast Cancer to Folate Restriction.Nutrients. 2021 May 13;13(5):1637. doi: 10.3390/nu13051637. Nutrients. 2021. PMID: 34068120 Free PMC article.
-
Targeting triple-negative breast cancer cells with the histone deacetylase inhibitor panobinostat.Breast Cancer Res. 2012 May 21;14(3):R79. doi: 10.1186/bcr3192. Breast Cancer Res. 2012. PMID: 22613095 Free PMC article.
-
Cell-Free DNA (cfDNA) Regulates Metabolic Remodeling, Sustaining Proliferation, Quiescence, and Migration in MDA-MB-231, a Triple-Negative Breast Carcinoma (TNBC) Cell Line.Metabolites. 2025 Mar 27;15(4):227. doi: 10.3390/metabo15040227. Metabolites. 2025. PMID: 40278356 Free PMC article.
Cited by
-
Therapeutic and diagnostic potential of extracellular vesicle (EV)-mediated intercellular transfer of mitochondria and mitochondrial components.J Cereb Blood Flow Metab. 2025 May 14:271678X251338971. doi: 10.1177/0271678X251338971. Online ahead of print. J Cereb Blood Flow Metab. 2025. PMID: 40367392 Free PMC article. Review.
-
Platelet Extracellular Vesicles as Natural Delivery Vehicles for Mitochondrial Dysfunction Therapy?ACS Biomater Sci Eng. 2025 May 12;11(5):2601-2621. doi: 10.1021/acsbiomaterials.5c00473. Epub 2025 Apr 25. ACS Biomater Sci Eng. 2025. PMID: 40280866 Free PMC article. Review.
-
Mitochondrial Antiviral Signaling Protein Activation by Retinoic Acid-Inducible Gene I Agonist Triggers Potent Antiviral Defense in Umbilical Cord Mesenchymal Stromal Cells Without Compromising Mitochondrial Function.Int J Mol Sci. 2025 May 14;26(10):4686. doi: 10.3390/ijms26104686. Int J Mol Sci. 2025. PMID: 40429828 Free PMC article.
-
Osteoblast-Derived Mitochondria Formulated with Cationic Liposome Guide Mesenchymal Stem Cells into Osteogenic Differentiation.Adv Sci (Weinh). 2025 Mar;12(12):e2412621. doi: 10.1002/advs.202412621. Epub 2025 Jan 31. Adv Sci (Weinh). 2025. PMID: 39887937 Free PMC article.
-
Mitochondria break free: Mitochondria-derived vesicles in aging and associated conditions.Ageing Res Rev. 2024 Dec;102:102549. doi: 10.1016/j.arr.2024.102549. Epub 2024 Oct 19. Ageing Res Rev. 2024. PMID: 39427885 Review.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

