Microbiota affects mitochondria and immune cell infiltrations via alternative polyadenylation during postnatal heart development
- PMID: 38283994
- PMCID: PMC10820713
- DOI: 10.3389/fcell.2023.1310409
Microbiota affects mitochondria and immune cell infiltrations via alternative polyadenylation during postnatal heart development
Abstract
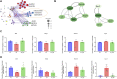
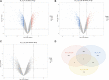
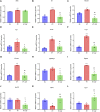
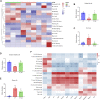
There is a growing body of evidence supporting the significant impact of microbiota on heart development. Alternative polyadenylation (APA) is a crucial mechanism for gene expression regulation and has been implicated in postnatal heart development. Nonetheless, whether microbiota can influence postnatal heart development through the regulation of APA remains unclear. Therefore, we conducted APA sequencing on heart tissues collected from specific pathogen-free (SPF) mice and germ-free (GF) mice at three different developmental stages: within the first 24 h after birth (P1), 7-day-old SPF mice, and 7-day-old GF mice. This approach allowed us to obtain a comprehensive genome-wide profile of APA sites in the heart tissue samples. In this study, we made a significant observation that GF mice exhibited noticeably longer 3' untranslated region (3' UTR) lengths. Furthermore, we confirmed significant alterations in the 3' UTR lengths of mitochondria-related genes, namely Rala, Timm13, and Uqcc3. Interestingly, the GF condition resulted in a marked decrease in mitochondrial cristae density and a reduction in the level of Tomm20 in postnatal hearts. Moreover, we discovered a connection between Rala and Src, which further implicated their association with other differentially expressed genes (DEGs). Notably, most of the DEGs were significantly downregulated in GF mice, with the exceptions being Thbs1 and Egr1. Importantly, the GF condition demonstrated a correlation with a lower infiltration of immune cells, whereby the levels of resting NK cells, Th17 cells, immature dendritic cells, and plasma cells in GF mice were comparable to those observed in P1 mice. Furthermore, we established significant correlations between these immune cells and Rala as well as the related DEGs. Our findings clearly indicated that microbiota plays a vital role in postnatal heart development by affecting APA switching, mitochondria and immune cell infiltrations.
Keywords: alternative polyadenylation; heart development; immune cell; microbiota; mitochondrion.
Copyright © 2024 Liu, Shao, Han, Zhu, Tu, Ma, Zhang, Yang and Chen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
LinkOut - more resources
Full Text Sources
Miscellaneous

