Abundance of G-Quadruplex Forming Sequences in the Hepatitis Delta Virus Genomes
- PMID: 38284014
- PMCID: PMC10809645
- DOI: 10.1021/acsomega.3c09288
Abundance of G-Quadruplex Forming Sequences in the Hepatitis Delta Virus Genomes
Abstract
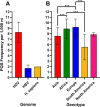
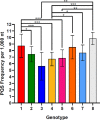
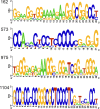
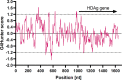
Hepatitis delta virus (HDV) is a highly unusual RNA satellite virus that depends on the presence of hepatitis B virus (HBV) to be infectious. Its compact and variable single-stranded RNA genome consists of eight major genotypes distributed unevenly across different continents. The significance of noncanonical secondary structures such as G-quadruplexes (G4s) is increasingly recognized at the DNA and RNA levels, particularly for transcription, replication, and translation. G4s are formed from guanine-rich sequences and have been identified in the vast majority of viral, eukaryotic, and prokaryotic genomes. In this study, we analyzed the G4 propensity of HDV genomes by using G4Hunter. Unlike HBV, which has a G4 density similar to that of the human genome, HDV displays a significantly higher number of potential quadruplex-forming sequences (PQS), with a density more than four times greater than that of the human genome. This finding suggests a critical role for G4s in HDV, especially given that the PQS regions are conserved across HDV genotypes. Furthermore, the prevalence of G4-forming sequences may represent a promising target for therapeutic interventions to control HDV replication.
© 2024 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
References
LinkOut - more resources
Full Text Sources
