Biochemical and Structural Analysis of the Bacterial Enzyme Succinyl-Diaminopimelate Desuccinylase (DapE) from Acinetobacter baumannii
- PMID: 38284080
- PMCID: PMC10809365
- DOI: 10.1021/acsomega.3c08231
Biochemical and Structural Analysis of the Bacterial Enzyme Succinyl-Diaminopimelate Desuccinylase (DapE) from Acinetobacter baumannii
Abstract
There is an urgent need for new antibiotics given the rise of antibiotic resistance, and succinyl-diaminopimelate desuccinylase (DapE, E.C. 3.5.1.18) has emerged as a promising bacterial enzyme target. DapE from Haemophilus influenzae (HiDapE) has been studied and inhibitors identified, but it is essential to explore DapE from different species to assess selective versus broad-spectrum therapeutics. We have determined the structure of DapE from the ESKAPE pathogen Acinetobacter baumannii (AbDapE) and studied inhibition by known inhibitors of HiDapE. AbDapE is inhibited by captopril and sulfate comparable to HiDapE, but AbDapE was not significantly inhibited by a known indoline sulfonamide HiDapE inhibitor. Captopril and sulfate both stabilize HiDapE by increasing the thermal melting temperature (Tm) in thermal shift assays. By contrast, sulfate decreases the stability of the AbDapE enzyme, whereas captopril increases the stability. Further, we report two crystal structures of selenomethionine-substituted AbDapE in the closed conformation, one with AbDapE in complex with succinate derived from enzymatic hydrolysis of N6-methyl-l,l-SDAP substrate and acetate (PDB code 7T1Q, 2.25 Å resolution), and a crystal structure of AbDapE with bound succinate along with l-(S)-lactate, a product of degradation of citric acid from the crystallization buffer during X-ray irradiation (PDB code 8F8O, 2.10 Å resolution).
© 2024 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
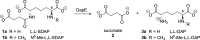
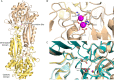
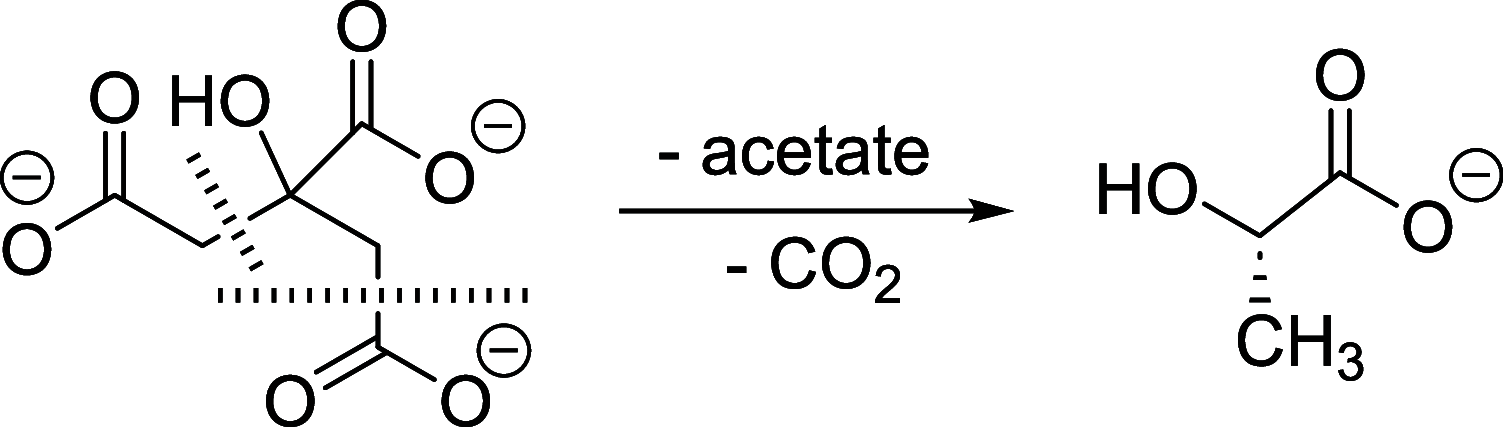

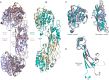
Similar articles
-
Synthesis and characterization of the N-succinyl-l,l-diaminopimelic acid desuccinylase (DapE) alternate substrate analog N,N-dimethyl-l,l-SDAP.Bioorg Med Chem. 2023 Aug 15;91:117415. doi: 10.1016/j.bmc.2023.117415. Epub 2023 Jul 12. Bioorg Med Chem. 2023. PMID: 37459673
-
Practical spectrophotometric assay for the dapE-encoded N-succinyl-L,L-diaminopimelic acid desuccinylase, a potential antibiotic target.PLoS One. 2018 Apr 26;13(4):e0196010. doi: 10.1371/journal.pone.0196010. eCollection 2018. PLoS One. 2018. PMID: 29698518 Free PMC article.
-
Inhibition of the dapE-Encoded N-Succinyl-L,L-diaminopimelic Acid Desuccinylase from Neisseria meningitidis by L-Captopril.Biochemistry. 2015 Aug 11;54(31):4834-44. doi: 10.1021/acs.biochem.5b00475. Epub 2015 Aug 3. Biochemistry. 2015. PMID: 26186504 Free PMC article.
-
Lysine biosynthesis in bacteria: a metallodesuccinylase as a potential antimicrobial target.J Biol Inorg Chem. 2013 Feb;18(2):155-163. doi: 10.1007/s00775-012-0965-1. Epub 2012 Dec 8. J Biol Inorg Chem. 2013. PMID: 23223968 Free PMC article. Review.
-
Deciphering the impact of Acinetobacter baumannii on human health, and exploration of natural compounds as efflux pump inhibitors to treat multidrug resistance.J Med Microbiol. 2024 Aug;73(8). doi: 10.1099/jmm.0.001867. J Med Microbiol. 2024. PMID: 39212030 Review.
Cited by
-
Reconstruction and Analysis of a Genome-Scale Metabolic Model of Acinetobacter lwoffii.Int J Mol Sci. 2024 Aug 28;25(17):9321. doi: 10.3390/ijms25179321. Int J Mol Sci. 2024. PMID: 39273268 Free PMC article.
-
N α -acetyl-L-ornithine deacetylase from Escherichia coli and a ninhydrin-based assay to enable inhibitor identification.Front Chem. 2024 Jul 11;12:1415644. doi: 10.3389/fchem.2024.1415644. eCollection 2024. Front Chem. 2024. PMID: 39055043 Free PMC article.
References
-
- Antibacterial Agents in Clinical Development: An Analysis of the Antibacterial Clinical Development Pipeline, Including Tuberculosis; World Health Organization, 2017.
Grants and funding
LinkOut - more resources
Full Text Sources
