A novel PANoptosis-related long non-coding RNA index to predict prognosis, immune microenvironment and personalised treatment in hepatocellular carcinoma
- PMID: 38284890
- PMCID: PMC10911344
- DOI: 10.18632/aging.205488
A novel PANoptosis-related long non-coding RNA index to predict prognosis, immune microenvironment and personalised treatment in hepatocellular carcinoma
Abstract
Background: PANoptosis is involved in the interaction of apoptosis, necroptosis and pyroptosis, playing a role in programmed cell death. Moreover, long non-coding RNAs (lncRNAs) regulate the PCD. This work aims to explore the role of PANoptosis-associated lncRNAs in hepatocellular carcinoma (HCC).
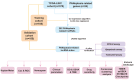
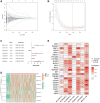
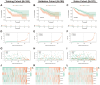
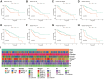
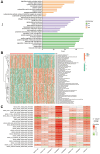
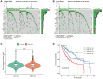
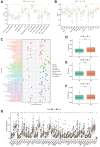
Methods: Co-expression analysis identified PANoptosis-associated lncRNAs in HCC. Cox and Least Absolute Shrinkage and Selection Operator (LASSO) algorithms were utilised to filter lncRNAs and establish a PANoptosis-related lncRNA index (PANRI). Additionally, Cox, Kaplan-Meier and receiver operating characteristic (ROC) curves were utilised to systematically evaluate the PANRI. Furthermore, Estimation of STromal and Immune cells in MAlignant Tumor tissues using Expression data (ESTIMATE), single sample gene set enrichment analysis (ssGSEA) and immune checkpoints were performed to analyse the potential of the PANRI in differentiating different tumour immune microenvironment (TIME) populations. The consensus clustering algorithm was used to distinguish individuals with HCC having different TIME subtypes. Finally, HCC cell lines HepG2 were utilised for further validation in in vitro experiments.
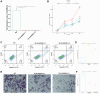
Results: The PANRI differentiates patients according to risk. Notably, ESTIMATE and ssGSEA algorithms revealed a high immune infiltration status in high-risk patients. Additionally, consensus clustering divided the patients into three clusters to identify different subtypes of TIME. Moreover, in vitro results showed that siRNA-mediated silencing of AL049840.4 inhibited the viability and migration of HepG2 cells and promoted apoptosis.
Conclusions: This is the first PANoptosis-related, lncRNA-based risk index in HCC to assess patient prognosis, TIME and response to immunotherapy. This study offers novel perspectives on the role of PANoptosis-associated lncRNAs in HCC.
Keywords: PANoptosis; hepatocellular carcinoma; lncRNA; prognosis; tumour immune microenvironment.
Conflict of interest statement
Figures




Similar articles
-
Identification of PANoptosis-related lncRNAs in hepatocellular carcinoma based on bioinformatics and construction of a prognostic model.Yi Chuan. 2025 Apr;47(4):456-475. doi: 10.16288/j.yczz.24-208. Yi Chuan. 2025. PMID: 40204520
-
Identification of molecular subtypes based on PANoptosis-related genes and construction of a signature for predicting the prognosis and response to immunotherapy response in hepatocellular carcinoma.Front Immunol. 2023 Aug 18;14:1218661. doi: 10.3389/fimmu.2023.1218661. eCollection 2023. Front Immunol. 2023. PMID: 37662906 Free PMC article.
-
A Cuproptosis-Related LncRNA Risk Model for Predicting Prognosis and Immunotherapeutic Efficacy in Patients with Hepatocellular Carcinoma.Biochem Genet. 2024 Jun;62(3):2332-2351. doi: 10.1007/s10528-023-10539-x. Epub 2023 Oct 29. Biochem Genet. 2024. PMID: 37898914
-
Long non-coding rnas as key modulators of the immune microenvironment in hepatocellular carcinoma: implications for Immunotherapy.Front Immunol. 2025 Apr 25;16:1523190. doi: 10.3389/fimmu.2025.1523190. eCollection 2025. Front Immunol. 2025. PMID: 40352941 Free PMC article. Review.
-
PANoptosis: bridging apoptosis, pyroptosis, and necroptosis in cancer progression and treatment.Cancer Gene Ther. 2024 Jul;31(7):970-983. doi: 10.1038/s41417-024-00765-9. Epub 2024 Mar 29. Cancer Gene Ther. 2024. PMID: 38553639 Free PMC article. Review.
Cited by
-
PANoptosis in cancer: bridging molecular mechanisms to therapeutic innovations.Cell Mol Immunol. 2025 Jul 28. doi: 10.1038/s41423-025-01329-z. Online ahead of print. Cell Mol Immunol. 2025. PMID: 40721869 Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

