STAT6 mutations enriched at diffuse large B-cell lymphoma relapse reshape the tumor microenvironment
- PMID: 38285120
- PMCID: PMC10920476
- DOI: 10.1007/s12185-023-03692-x
STAT6 mutations enriched at diffuse large B-cell lymphoma relapse reshape the tumor microenvironment
Abstract
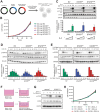
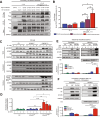
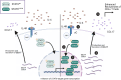
Diffuse large B-cell lymphoma (DLBCL) relapses in approximately 40% of patients following frontline therapy. We reported that STAT6D419 mutations are enriched in relapsed/refractory DLBCL (rrDLBCL) samples, suggesting that JAK/STAT signaling plays a role in therapeutic resistance. We hypothesized that STAT6D419 mutations can improve DLBCL cell survival by reprogramming the microenvironment to sustain STAT6 activation. Thus, we investigated the role of STAT6D419 mutations on DLBCL cell growth and its microenvironment. We found that phospho-STAT6D419N was retained in the nucleus longer than phospho-STAT6WT following IL-4 stimulation, and STAT6D419N recognized a more restricted DNA-consensus sequence than STAT6WT. Upon IL-4 induction, STAT6D419N expression led to a higher magnitude of gene expression changes, but in a more selective list of gene targets compared with STATWT. The most significantly expressed genes induced by STAT6D419N were those implicated in survival, proliferation, migration, and chemotaxis, in particular CCL17. This chemokine, also known as TARC, attracts helper T-cells to the tumor microenvironment, especially in Hodgkin's lymphoma. To this end, in DLBCL, phospho-STAT6+ rrDLBCL cells had a greater proportion of infiltrating CD4+ T-cells than phospho-STAT6- tumors. Our findings suggest that STAT6D419 mutations in DLBCL lead to cell autonomous changes, enhanced signaling, and altered composition of the tumor microenvironment.
Keywords: DLBCL; STAT6; Tumor microenvironment.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no potential conflicts of interest.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

