Glioblastoma and radiotherapy: A multicenter AI study for Survival Predictions from MRI (GRASP study)
- PMID: 38285679
- PMCID: PMC11145448
- DOI: 10.1093/neuonc/noae017
Glioblastoma and radiotherapy: A multicenter AI study for Survival Predictions from MRI (GRASP study)
Abstract
Background: The aim was to predict survival of glioblastoma at 8 months after radiotherapy (a period allowing for completing a typical course of adjuvant temozolomide), by applying deep learning to the first brain MRI after radiotherapy completion.
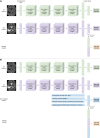
Methods: Retrospective and prospective data were collected from 206 consecutive glioblastoma, isocitrate dehydrogenase -wildtype patients diagnosed between March 2014 and February 2022 across 11 UK centers. Models were trained on 158 retrospective patients from 3 centers. Holdout test sets were retrospective (n = 19; internal validation), and prospective (n = 29; external validation from 8 distinct centers). Neural network branches for T2-weighted and contrast-enhanced T1-weighted inputs were concatenated to predict survival. A nonimaging branch (demographics/MGMT/treatment data) was also combined with the imaging model. We investigated the influence of individual MR sequences; nonimaging features; and weighted dense blocks pretrained for abnormality detection.
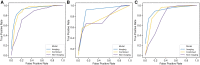
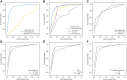
Results: The imaging model outperformed the nonimaging model in all test sets (area under the receiver-operating characteristic curve, AUC P = .038) and performed similarly to a combined imaging/nonimaging model (P > .05). Imaging, nonimaging, and combined models applied to amalgamated test sets gave AUCs of 0.93, 0.79, and 0.91. Initializing the imaging model with pretrained weights from 10 000s of brain MRIs improved performance considerably (amalgamated test sets without pretraining 0.64; P = .003).
Conclusions: A deep learning model using MRI images after radiotherapy reliably and accurately determined survival of glioblastoma. The model serves as a prognostic biomarker identifying patients who will not survive beyond a typical course of adjuvant temozolomide, thereby stratifying patients into those who might require early second-line or clinical trial treatment.
Keywords: artificial intelligence; deep learning; glioblastoma; magnetic resonance imaging; survival.
© The Author(s) 2024. Published by Oxford University Press on behalf of the Society for Neuro-Oncology.
Conflict of interest statement
There is no conflict of interest for all authors as a consortium. A.C. - None declared. D.A.W—None declared. L.S.C.—None declared. H.S.—None declared. S.C.—None declared. K.Fa.—None declared. R.F.—None declared. C.R-H.—None declared. S.Th.—None declared. S.J.W.—None declared. S.Te.—None declared. C.M. -None declared. K.Fo.—None declared. M.W.—stock and other interests: PearBio. Q.W.—None declared. A.R.—None declared. C.D.—None declared. M.Ma.—None declared. Y.H.L.—None declared. C.A.L.—Nonedeclared. A.B.—None declared. A.L.—None declared. T.Y.—None declared. J.B.—None declared. E.C.—None declared. E.B.—None declared. T.-C.L.—None declared. L.W.—None declared. J.L.—None declared. R.M.—consultancy: Brainlab, Stryker; payment/honoraria: Baxter, Roswell Comprehensive Cancer Centre, Zeiss; support for attending meetings/travel: Brainlab, Roswell Comprehensive Cancer Centre, Zeiss; patents: UK patent office; unpaid leadership/fiduciary role: Oscar’s Paediatric Brain Tumour Charity, TJBCM-BTR NTA; shareholding: Opto Biosystems, RBM Healthcare, Assemblify; clinical advisor: MHRA. E.K.—None declared. R.B.—None declared. D.B.—None declared. J.G.—None declared. L.B.—None declared. A.S.—None declared. K.A.—None declared. S.O.—consultancy: Proximie, Avatera Medical; stock: Hypervision Surgical Ltd. M.Mo.—None declared. T.C.B.—consultancy: Microvention; payment/honoraria for education lectures: Siemens Healthineers Speakers Bureau, Medtronic Speakers Bureau; support for attending meetings/travel: Balt.
Figures

References
-
- Brodbelt A, Greenberg D, Winters T, et al.; (UK) National Cancer Information Network Brain Tumour Group. Glioblastoma in England: 2007–2011. Eur J Cancer. 2015;51(4):533–542. - PubMed
-
- Weller M, Van Den Bent M, Tonn JC, et al.; European Association for Neuro-Oncology (EANO) Task Force on Gliomas. European Association for Neuro-Oncology (EANO) guideline on the diagnosis and treatment of adult astrocytic and oligodendroglial gliomas. Lancet Oncol. 2017;18(6):e315–e329. - PubMed
-
- Fu X, Chen C, Li D.. Survival prediction of patients suffering from glioblastoma based on two-branch DenseNet using multi-channel features. Int J Comput Assist Radiol Surg. 2021;16(2):207–217. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- PRP/067/20Fx/Innovation and Technology Commission
- Leeds Hospitals Charity
- Novocure
- Yorkshire's Brain Tumour Charity
- MR/W021684/1/MRC_/Medical Research Council/United Kingdom
- 203914/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- 28832/CRUK_/Cancer Research UK/United Kingdom
- MR/N013700/1/MRC_/Medical Research Council/United Kingdom
- Candlelighters
- National Institute for Health and Care Research Imperial Biomedical Research Centre
- Macmillan Cancer Care
- National Institute for Health
- EP/T022205/1/EPSRC
- RCCCTF-OCT22/100002/CRUK_/Cancer Research UK/United Kingdom
- 215010/Z/18/Z/WT_/Wellcome Trust/United Kingdom
- Brain Tumour Charity
- King's College London
- 203148/Z/16/Z/Wellcome EPSRC Centre for Medical Engineering
- WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Medical
Research Materials

