Proteomic Biomarkers of Quantitative Interstitial Abnormalities in COPDGene and CARDIA Lung Study
- PMID: 38285918
- PMCID: PMC11092953
- DOI: 10.1164/rccm.202307-1129OC
Proteomic Biomarkers of Quantitative Interstitial Abnormalities in COPDGene and CARDIA Lung Study
Abstract
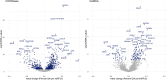
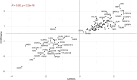
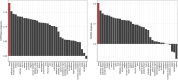
Rationale: Quantitative interstitial abnormalities (QIAs) are early measures of lung injury automatically detected on chest computed tomography scans. QIAs are associated with impaired respiratory health and share features with advanced lung diseases, but their biological underpinnings are not well understood. Objectives: To identify novel protein biomarkers of QIAs using high-throughput plasma proteomic panels within two multicenter cohorts. Methods: We measured the plasma proteomics of 4,383 participants in an older, ever-smoker cohort (COPDGene [Genetic Epidemiology of Chronic Obstructive Pulmonary Disease]) and 2,925 participants in a younger population cohort (CARDIA [Coronary Artery Disease Risk in Young Adults]) using the SomaLogic SomaScan assays. We measured QIAs using a local density histogram method. We assessed the associations between proteomic biomarker concentrations and QIAs using multivariable linear regression models adjusted for age, sex, body mass index, smoking status, and study center (Benjamini-Hochberg false discovery rate-corrected P ⩽ 0.05). Measurements and Main Results: In total, 852 proteins were significantly associated with QIAs in COPDGene and 185 in CARDIA. Of the 144 proteins that overlapped between COPDGene and CARDIA, all but one shared directionalities and magnitudes. These proteins were enriched for 49 Gene Ontology pathways, including biological processes in inflammatory response, cell adhesion, immune response, ERK1/2 regulation, and signaling; cellular components in extracellular regions; and molecular functions including calcium ion and heparin binding. Conclusions: We identified the proteomic biomarkers of QIAs in an older, smoking population with a higher prevalence of pulmonary disease and in a younger, healthier community cohort. These proteomics features may be markers of early precursors of advanced lung diseases.
Keywords: biomarkers; interstitial lung disease; proteomics; pulmonary emphysema.
Figures

Comment in
-
Mapping the Proteomic Landscape of Radiological Lung Abnormalities.Am J Respir Crit Care Med. 2024 May 1;209(9):1052-1054. doi: 10.1164/rccm.202402-0310ED. Am J Respir Crit Care Med. 2024. PMID: 38442249 Free PMC article. No abstract available.
Similar articles
-
The Proteomic Profile of Interstitial Lung Abnormalities.Am J Respir Crit Care Med. 2022 Aug 1;206(3):337-346. doi: 10.1164/rccm.202110-2296OC. Am J Respir Crit Care Med. 2022. PMID: 35438610 Free PMC article.
-
Plasma metabolomics and quantitative interstitial abnormalities in ever-smokers.Respir Res. 2023 Nov 4;24(1):265. doi: 10.1186/s12931-023-02576-2. Respir Res. 2023. PMID: 37925418 Free PMC article.
-
Omics and the Search for Blood Biomarkers in Chronic Obstructive Pulmonary Disease. Insights from COPDGene.Am J Respir Cell Mol Biol. 2019 Aug;61(2):143-149. doi: 10.1165/rcmb.2018-0245PS. Am J Respir Cell Mol Biol. 2019. PMID: 30874442 Free PMC article. Review.
-
Blood-based Transcriptomic and Proteomic Biomarkers of Emphysema.Am J Respir Crit Care Med. 2024 Feb 1;209(3):273-287. doi: 10.1164/rccm.202301-0067OC. Am J Respir Crit Care Med. 2024. PMID: 37917913 Free PMC article.
-
Imaging Advances in Chronic Obstructive Pulmonary Disease. Insights from the Genetic Epidemiology of Chronic Obstructive Pulmonary Disease (COPDGene) Study.Am J Respir Crit Care Med. 2019 Feb 1;199(3):286-301. doi: 10.1164/rccm.201807-1351SO. Am J Respir Crit Care Med. 2019. PMID: 30304637 Free PMC article. Review.
Cited by
-
Are We Getting Closer to the "Cholesterol" for Chronic Respiratory Disease?Am J Respir Crit Care Med. 2024 Sep 12;211(1):6-7. doi: 10.1164/rccm.202407-1480ED. Online ahead of print. Am J Respir Crit Care Med. 2024. PMID: 39265184 Free PMC article. No abstract available.
-
Approach to the Evaluation and Management of Interstitial Lung Abnormalities: An Official American Thoracic Society Clinical Statement.Am J Respir Crit Care Med. 2025 Jul;211(7):1132-1155. doi: 10.1164/rccm.202505-1054ST. Am J Respir Crit Care Med. 2025. PMID: 40387336 Free PMC article. Review.
-
Protein biomarkers of interstitial lung abnormalities in relatives of patients with pulmonary fibrosis.Eur Respir J. 2025 Jun 5;65(6):2401349. doi: 10.1183/13993003.01349-2024. Print 2025 Jun. Eur Respir J. 2025. PMID: 39884764 Free PMC article.
-
Plasma proteomic analysis of intermuscular fat links muscle integrity with processing speed in older adults.Alzheimers Dement. 2025 May;21(5):e70261. doi: 10.1002/alz.70261. Alzheimers Dement. 2025. PMID: 40390202 Free PMC article.
-
Plasma proteomic analysis of intermuscular fat links muscle integrity with processing speed in older adults.medRxiv [Preprint]. 2025 Jan 27:2025.01.24.25320976. doi: 10.1101/2025.01.24.25320976. medRxiv. 2025. Update in: Alzheimers Dement. 2025 May;21(5):e70261. doi: 10.1002/alz.70261. PMID: 39974123 Free PMC article. Updated. Preprint.
References
-
- Choi B, Adan N, Doyle TJ, San José Estépar R, Harmouche R, Humphries SM, et al. COPDGene Study and Pittsburgh Lung Screening Study Investigators Quantitative interstitial abnormality progression and outcomes in the Genetic Epidemiology of COPD and Pittsburgh Lung Screening Study cohorts. Chest . 2023;163:164–175. - PMC - PubMed
-
- King TE, Jr, Bradford WZ, Castro-Bernardini S, Fagan EA, Glaspole I, Glassberg MK, et al. ASCEND Study Group A phase 3 trial of pirfenidone in patients with idiopathic pulmonary fibrosis. N Engl J Med . 2014;370:2083–2092. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 HL137995/HL/NHLBI NIH HHS/United States
- HHSN268201800004I/HL/NHLBI NIH HHS/United States
- U01 HL089897/HL/NHLBI NIH HHS/United States
- R01 HL116931/HL/NHLBI NIH HHS/United States
- HHSN268201800003I/HL/NHLBI NIH HHS/United States
- HHSN268201800007I/HL/NHLBI NIH HHS/United States
- F32 HL167486/HL/NHLBI NIH HHS/United States
- HHSN268201800005I/HL/NHLBI NIH HHS/United States
- P01 HL114501/HL/NHLBI NIH HHS/United States
- R21 HL140422/HL/NHLBI NIH HHS/United States
- R01 HL145372/HL/NHLBI NIH HHS/United States
- R21 LM013670/LM/NLM NIH HHS/United States
- U01 HL089856/HL/NHLBI NIH HHS/United States
- R01 HL122477/HL/NHLBI NIH HHS/United States
- HHSN268201800006I/HL/NHLBI NIH HHS/United States
- F32HL167486/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

