COPD basal cells are primed towards secretory to multiciliated cell imbalance driving increased resilience to environmental stressors
- PMID: 38286613
- PMCID: PMC11137452
- DOI: 10.1136/thorax-2022-219958
COPD basal cells are primed towards secretory to multiciliated cell imbalance driving increased resilience to environmental stressors
Abstract
Introduction: Environmental pollutants injure the mucociliary elevator, thereby provoking disease progression in chronic obstructive pulmonary disease (COPD). Epithelial resilience mechanisms to environmental nanoparticles in health and disease are poorly characterised.
Methods: We delineated the impact of prevalent pollutants such as carbon and zinc oxide nanoparticles, on cellular function and progeny in primary human bronchial epithelial cells (pHBECs) from end-stage COPD (COPD-IV, n=4), early disease (COPD-II, n=3) and pulmonary healthy individuals (n=4). After nanoparticle exposure of pHBECs at air-liquid interface, cell cultures were characterised by functional assays, transcriptome and protein analysis, complemented by single-cell analysis in serial samples of pHBEC cultures focusing on basal cell differentiation.
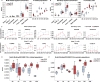
Results: COPD-IV was characterised by a prosecretory phenotype (twofold increase in MUC5AC+) at the expense of the multiciliated epithelium (threefold reduction in Ac-Tub+), resulting in an increased resilience towards particle-induced cell damage (fivefold reduction in transepithelial electrical resistance), as exemplified by environmentally abundant doses of zinc oxide nanoparticles. Exposure of COPD-II cultures to cigarette smoke extract provoked the COPD-IV characteristic, prosecretory phenotype. Time-resolved single-cell transcriptomics revealed an underlying COPD-IV unique basal cell state characterised by a twofold increase in KRT5+ (P=0.018) and LAMB3+ (P=0.050) expression, as well as a significant activation of Wnt-specific (P=0.014) and Notch-specific (P=0.021) genes, especially in precursors of suprabasal and secretory cells.
Conclusion: We identified COPD stage-specific gene alterations in basal cells that affect the cellular composition of the bronchial elevator and may control disease-specific epithelial resilience mechanisms in response to environmental nanoparticles. The identified phenomena likely inform treatment and prevention strategies.
Keywords: Airway Epithelium; COPD Pathology; COPD exacerbations mechanisms; Occupational Lung Disease; Thoracic Surgery.
© Author(s) (or their employer(s)) 2024. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: None declared.
Figures





References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous
