Narrowing down the candidates of beneficial A-to-I RNA editing by comparing the recoding sites with uneditable counterparts
- PMID: 38286757
- PMCID: PMC10826634
- DOI: 10.1080/19491034.2024.2304503
Narrowing down the candidates of beneficial A-to-I RNA editing by comparing the recoding sites with uneditable counterparts
Abstract
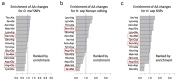
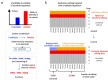
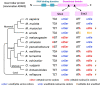
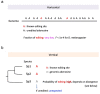
Adar-mediated adenosine-to-inosine (A-to-I) RNA editing mainly occurs in nucleus and diversifies the transcriptome in a flexible manner. It has been a challenging task to identify beneficial editing sites from the sea of total editing events. The functional Ser>Gly auto-recoding site in insect Adar gene has uneditable Ser codons in ancestral nodes, indicating the selective advantage to having an editable status. Here, we extended this case study to more metazoan species, and also looked for all Drosophila recoding events with potential uneditable synonymous codons. Interestingly, in D. melanogaster, the abundant nonsynonymous editing is enriched in the codons that have uneditable counterparts, but the Adar Ser>Gly case suggests that the editable orthologous codons in other species are not necessarily edited. The use of editable versus ancestral uneditable codon is a smart way to infer the selective advantage of RNA editing, and priority might be given to these editing sites for functional studies due to the feasibility to construct an uneditable allele. Our study proposes an idea to narrow down the candidates of beneficial recoding sites. Meanwhile, we stress that the matched transcriptomes are needed to verify the conservation of editing events during evolution.
Keywords: A-to-I RNA editing; Adar; auto-recoding; beneficial; editable; metazoan.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
