Location-agnostic site-specific protein bioconjugation via Baylis Hillman adducts
- PMID: 38286847
- PMCID: PMC10825175
- DOI: 10.1038/s41467-024-45124-2
Location-agnostic site-specific protein bioconjugation via Baylis Hillman adducts
Abstract
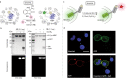
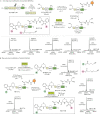
Proteins labelled site-specifically with small molecules are valuable assets for chemical biology and drug development. The unique reactivity profile of the 1,2-aminothiol moiety of N-terminal cysteines (N-Cys) of proteins renders it highly attractive for regioselective protein labelling. Herein, we report an ultrafast Z-selective reaction between isatin-derived Baylis Hillman adducts and 1,2-aminothiols to form a bis-heterocyclic scaffold, and employ it for stable protein bioconjugation under both in vitro and live-cell conditions. We refer to our protein bioconjugation technology as Baylis Hillman orchestrated protein aminothiol labelling (BHoPAL). Furthermore, we report a lipoic acid ligase-based technology for introducing the 1,2-aminothiol moiety at any desired site within proteins, rendering BHoPAL location-agnostic (not limited to N-Cys). By using this approach in tandem with BHoPAL, we generate dually labelled protein bioconjugates appended with different labels at two distinct specific sites on a single protein molecule. Taken together, the protein bioconjugation toolkit that we disclose herein will contribute towards the generation of both mono and multi-labelled protein-small molecule bioconjugates for applications as diverse as biophysical assays, cellular imaging, and the production of therapeutic protein-drug conjugates. In addition to protein bioconjugation, the bis-heterocyclic scaffold we report herein will find applications in synthetic and medicinal chemistry.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Hoyt EA, Cal PMSD, Oliveira BL, Bernardes GJL. Contemporary approaches to site-selective protein modification. Nat. Rev. Chem. 2019;3:147–171. doi: 10.1038/s41570-019-0079-1. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

