A universal metabolite repair enzyme removes a strong inhibitor of the TCA cycle
- PMID: 38287013
- PMCID: PMC10825186
- DOI: 10.1038/s41467-024-45134-0
A universal metabolite repair enzyme removes a strong inhibitor of the TCA cycle
Abstract
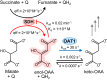
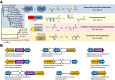
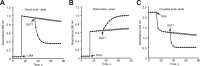
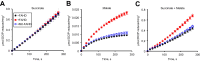
A prevalent side-reaction of succinate dehydrogenase oxidizes malate to enol-oxaloacetate (OAA), a metabolically inactive form of OAA that is a strong inhibitor of succinate dehydrogenase. We purified from cow heart mitochondria an enzyme (OAT1) with OAA tautomerase (OAT) activity that converts enol-OAA to the physiological keto-OAA form, and determined that it belongs to the highly conserved and previously uncharacterized Fumarylacetoacetate_hydrolase_domain-containing protein family. From all three domains of life, heterologously expressed proteins were shown to have strong OAT activity, and ablating the OAT1 homolog caused significant growth defects. In Escherichia coli, expression of succinate dehydrogenase was necessary for OAT1-associated growth defects to occur, and ablating OAT1 caused a significant increase in acetate and other metabolites associated with anaerobic respiration. OAT1 increased the succinate dehydrogenase reaction rate by 35% in in vitro assays with physiological concentrations of both succinate and malate. Our results suggest that OAT1 is a universal metabolite repair enzyme that is required to maximize aerobic respiration efficiency by preventing succinate dehydrogenase inhibition.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Direct demonstration of enol-oxaloacetate as an immediate product of malate oxidation by the mammalian succinate dehydrogenase.FEBS Lett. 1991 Jul 29;286(1-2):76-8. doi: 10.1016/0014-5793(91)80944-x. FEBS Lett. 1991. PMID: 1864383
-
Oxidation of malate by the mitochondrial succinate-ubiquinone reductase.Biochim Biophys Acta. 1988 Oct 26;936(1):1-9. doi: 10.1016/0005-2728(88)90245-9. Biochim Biophys Acta. 1988. PMID: 2902878
-
Metabolic clearance of oxaloacetate and mitochondrial complex II respiration: Divergent control in skeletal muscle and brown adipose tissue.Biochim Biophys Acta Bioenerg. 2023 Jan 1;1864(1):148930. doi: 10.1016/j.bbabio.2022.148930. Epub 2022 Oct 19. Biochim Biophys Acta Bioenerg. 2023. PMID: 36272463 Free PMC article.
-
Metabolic regulatory functions of oxalacetate.Horiz Biochem Biophys. 1974;1:175-219. Horiz Biochem Biophys. 1974. PMID: 4619614 Review. No abstract available.
-
Enzyme promiscuity, metabolite damage, and metabolite damage control systems of the tricarboxylic acid cycle.FEBS J. 2020 Apr;287(7):1343-1358. doi: 10.1111/febs.15284. Epub 2020 Mar 18. FEBS J. 2020. PMID: 32149453 Review.
Cited by
-
Identification of age-associated microbial changes via long-read 16S sequencing.Gut Pathog. 2024 Oct 5;16(1):56. doi: 10.1186/s13099-024-00650-8. Gut Pathog. 2024. PMID: 39369250 Free PMC article.
-
Biochemical and genomic evidence for converging metabolic routes of metformin and biguanide breakdown in environmental Pseudomonads.J Biol Chem. 2024 Dec;300(12):107935. doi: 10.1016/j.jbc.2024.107935. Epub 2024 Oct 28. J Biol Chem. 2024. PMID: 39476966 Free PMC article.
-
FAHD1 and mitochondrial metabolism: a decade of pioneering discoveries.FEBS J. 2025 Jun;292(12):2973-2991. doi: 10.1111/febs.17345. Epub 2024 Dec 6. FEBS J. 2025. PMID: 39642098 Free PMC article. Review.
-
α-Amino-β-carboxymuconate-ε-semialdehyde decarboxylase catalyzes enol/keto tautomerization of oxaloacetate.J Biol Chem. 2024 Nov;300(11):107878. doi: 10.1016/j.jbc.2024.107878. Epub 2024 Oct 11. J Biol Chem. 2024. PMID: 39395800 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

