N1-methylation of adenosine (m1A) in ND5 mRNA leads to complex I dysfunction in Alzheimer's disease
- PMID: 38287100
- PMCID: PMC11189808
- DOI: 10.1038/s41380-024-02421-y
N1-methylation of adenosine (m1A) in ND5 mRNA leads to complex I dysfunction in Alzheimer's disease
Abstract
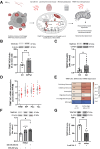
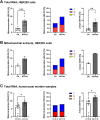
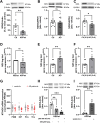
One mechanism of particular interest to regulate mRNA fate post-transcriptionally is mRNA modification. Especially the extent of m1A mRNA methylation is highly discussed due to methodological differences. However, one single m1A site in mitochondrial ND5 mRNA was unanimously reported by different groups. ND5 is a subunit of complex I of the respiratory chain. It is considered essential for the coupling of oxidation and proton transport. Here we demonstrate that this m1A site might be involved in the pathophysiology of Alzheimer's disease (AD). One of the pathological hallmarks of this neurodegenerative disease is mitochondrial dysfunction, mainly induced by Amyloid β (Aβ). Aβ mainly disturbs functions of complex I and IV of the respiratory chain. However, the molecular mechanism of complex I dysfunction is still not fully understood. We found enhanced m1A methylation of ND5 mRNA in an AD cell model as well as in AD patients. Formation of this m1A methylation is catalyzed by increased TRMT10C protein levels, leading to translation repression of ND5. As a consequence, here demonstrated for the first time, TRMT10C induced m1A methylation of ND5 mRNA leads to mitochondrial dysfunction. Our findings suggest that this newly identified mechanism might be involved in Aβ-induced mitochondrial dysfunction.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
The m1A landscape on cytosolic and mitochondrial mRNA at single-base resolution.Nature. 2017 Nov 9;551(7679):251-255. doi: 10.1038/nature24456. Epub 2017 Oct 25. Nature. 2017. PMID: 29072297
-
Impaired Cellular Bioenergetics Causes Mitochondrial Calcium Handling Defects in MT-ND5 Mutant Cybrids.PLoS One. 2016 Apr 25;11(4):e0154371. doi: 10.1371/journal.pone.0154371. eCollection 2016. PLoS One. 2016. PMID: 27110715 Free PMC article.
-
Impact of Amyloid-β on Platelet Mitochondrial Function and Platelet-Mediated Amyloid Aggregation in Alzheimer's Disease.Int J Mol Sci. 2021 Sep 6;22(17):9633. doi: 10.3390/ijms22179633. Int J Mol Sci. 2021. PMID: 34502546 Free PMC article.
-
Olesoxime improves cerebral mitochondrial dysfunction and enhances Aβ levels in preclinical models of Alzheimer's disease.Exp Neurol. 2020 Jul;329:113286. doi: 10.1016/j.expneurol.2020.113286. Epub 2020 Mar 18. Exp Neurol. 2020. PMID: 32199815 Review.
-
Mitochondria and Alzheimer's disease: amyloid-beta peptide uptake and degradation by the presequence protease, hPreP.J Bioenerg Biomembr. 2009 Oct;41(5):447-51. doi: 10.1007/s10863-009-9244-4. J Bioenerg Biomembr. 2009. PMID: 19798557 Review.
Cited by
-
Mitochondrial One-Carbon Metabolism and Alzheimer's Disease.Int J Mol Sci. 2024 Jun 7;25(12):6302. doi: 10.3390/ijms25126302. Int J Mol Sci. 2024. PMID: 38928008 Free PMC article. Review.
-
Feedback regulation between histone lactylation and ALKBH3-mediated glycolysis regulates age-related macular degeneration pathology.Proc Natl Acad Sci U S A. 2025 Jun 17;122(24):e2416046122. doi: 10.1073/pnas.2416046122. Epub 2025 Jun 10. Proc Natl Acad Sci U S A. 2025. PMID: 40493193
-
The RMaP challenge of predicting RNA modifications by nanopore sequencing.Commun Chem. 2025 Apr 12;8(1):115. doi: 10.1038/s42004-025-01507-0. Commun Chem. 2025. PMID: 40221591 Free PMC article.
-
ModiDeC: a multi-RNA modification classifier for direct nanopore sequencing.Nucleic Acids Res. 2025 Jul 19;53(14):gkaf673. doi: 10.1093/nar/gkaf673. Nucleic Acids Res. 2025. PMID: 40682823 Free PMC article.
-
Analysis of human brain RNA-seq data reveals combined effects of 4 types of RNA modifications and 18 types of programmed cell death on Alzheimer's disease.J Transl Med. 2025 Apr 3;23(1):396. doi: 10.1186/s12967-025-06324-6. J Transl Med. 2025. PMID: 40181382 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

