Optineurin provides a mitophagy contact site for TBK1 activation
- PMID: 38287189
- PMCID: PMC10907724
- DOI: 10.1038/s44318-024-00036-1
Optineurin provides a mitophagy contact site for TBK1 activation
Abstract
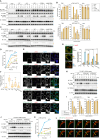
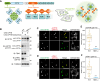
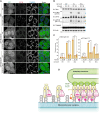
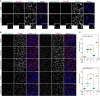
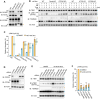
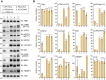
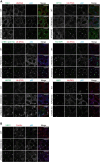
Tank-binding kinase 1 (TBK1) is a Ser/Thr kinase that is involved in many intracellular processes, such as innate immunity, cell cycle, and apoptosis. TBK1 is also important for phosphorylating the autophagy adaptors that mediate the selective autophagic removal of damaged mitochondria. However, the mechanism by which PINK1-Parkin-mediated mitophagy activates TBK1 remains largely unknown. Here, we show that the autophagy adaptor optineurin (OPTN) provides a unique platform for TBK1 activation. Both the OPTN-ubiquitin and the OPTN-pre-autophagosomal structure (PAS) interaction axes facilitate assembly of the OPTN-TBK1 complex at a contact sites between damaged mitochondria and the autophagosome formation sites. At this assembly point, a positive feedback loop for TBK1 activation is initiated that accelerates hetero-autophosphorylation of the protein. Expression of monobodies engineered here to bind OPTN impaired OPTN accumulation at contact sites, as well as the subsequent activation of TBK1, thereby inhibiting mitochondrial degradation. Taken together, these data show that a positive and reciprocal relationship between OPTN and TBK1 initiates autophagosome biogenesis on damaged mitochondria.
Keywords: Autophagy; Mitochondria; PINK1; Parkin; Ubiquitin.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

