Migrasomal autophagosomes relieve endoplasmic reticulum stress in glioblastoma cells
- PMID: 38287397
- PMCID: PMC10826056
- DOI: 10.1186/s12915-024-01829-w
Migrasomal autophagosomes relieve endoplasmic reticulum stress in glioblastoma cells
Abstract
Background: Glioblastoma (GBM) is more difficult to treat than other intractable adult tumors. The main reason that GBM is so difficult to treat is that it is highly infiltrative. Migrasomes are newly discovered membrane structures observed in migrating cells. Thus, they can be generated from GBM cells that have the ability to migrate along the brain parenchyma. However, the function of migrasomes has not yet been elucidated in GBM cells.
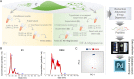
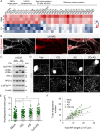
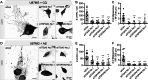
Results: Here, we describe the composition and function of migrasomes generated along with GBM cell migration. Proteomic analysis revealed that LC3B-positive autophagosomes were abundant in the migrasomes of GBM cells. An increased number of migrasomes was observed following treatment with chloroquine (CQ) or inhibition of the expression of STX17 and SNAP29, which are involved in autophagosome/lysosome fusion. Furthermore, depletion of ITGA5 or TSPAN4 did not relieve endoplasmic reticulum (ER) stress in cells, resulting in cell death.
Conclusions: Taken together, our study suggests that increasing the number of autophagosomes, through inhibition of autophagosome/lysosome fusion, generates migrasomes that have the capacity to alleviate cellular stress.
Keywords: Autophagosome; Cell death; ER stress; ITGA5; Migrasome; Retraction fiber; TSPAN4.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10(5):459–466. doi: 10.1016/S1470-2045(09)70025-7. - DOI - PubMed
Publication types
MeSH terms
Grants and funding
- 2015R1A5A1009024/National Research Foundation of Korea
- 2015R1A5A1009024/National Research Foundation of Korea
- 2019M3E5D3073106/National Research Foundation of Korea
- 2015R1A5A1009024/National Research Foundation of Korea
- RS-2023-00218838/Innovative Research Group Project of the National Natural Science Foundation of China
LinkOut - more resources
Full Text Sources
Miscellaneous

