Reprogramming of glucose metabolism: Metabolic alterations in the progression of osteosarcoma
- PMID: 38288377
- PMCID: PMC10823108
- DOI: 10.1016/j.jbo.2024.100521
Reprogramming of glucose metabolism: Metabolic alterations in the progression of osteosarcoma
Abstract
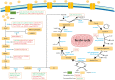
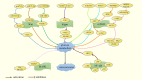
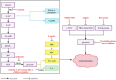
Metabolic reprogramming is an adaptive response of tumour cells under hypoxia and low nutrition conditions. There is increasing evidence that glucose metabolism reprogramming can regulate the growth and metastasis of osteosarcoma (OS). Reprogramming in the progress of OS can bring opportunities for early diagnosis and treatment of OS. Previous research mainly focused on the glycolytic pathway of glucose metabolism, often neglecting the tricarboxylic acid cycle and pentose phosphate pathway. However, the tricarboxylic acid cycle and pentose phosphate pathway of glucose metabolism are also involved in the progression of OS and are closely related to this disease. The research on glucose metabolism in OS has not yet been summarized. In this review, we discuss the abnormal expression of key molecules related to glucose metabolism in OS and summarize the glucose metabolism related signaling pathways involved in the occurrence and development of OS. In addition, we discuss some of the targeted drugs that regulate glucose metabolism pathways, which can lead to effective strategies for targeted treatment of OS.
Keywords: Glucose metabolism; Key enzymes; Osteosarcoma; Signaling pathways; Targeted therapy.
© 2024 The Authors. Published by Elsevier GmbH.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


Similar articles
-
Metabolic reprogramming of glucose: the metabolic basis for the occurrence and development of hepatocellular carcinoma.Front Oncol. 2025 Feb 6;15:1545086. doi: 10.3389/fonc.2025.1545086. eCollection 2025. Front Oncol. 2025. PMID: 39980550 Free PMC article. Review.
-
The roles of glycolysis in osteosarcoma.Front Pharmacol. 2022 Aug 17;13:950886. doi: 10.3389/fphar.2022.950886. eCollection 2022. Front Pharmacol. 2022. PMID: 36059961 Free PMC article. Review.
-
Metabolic dysregulation and emerging therapeutical targets for hepatocellular carcinoma.Acta Pharm Sin B. 2022 Feb;12(2):558-580. doi: 10.1016/j.apsb.2021.09.019. Epub 2021 Sep 25. Acta Pharm Sin B. 2022. PMID: 35256934 Free PMC article. Review.
-
P2RX7 promotes osteosarcoma progression and glucose metabolism by enhancing c-Myc stabilization.J Transl Med. 2023 Feb 20;21(1):132. doi: 10.1186/s12967-023-03985-z. J Transl Med. 2023. PMID: 36803784 Free PMC article.
-
Energy metabolism pathways in breast cancer progression: The reprogramming, crosstalk, and potential therapeutic targets.Transl Oncol. 2022 Dec;26:101534. doi: 10.1016/j.tranon.2022.101534. Epub 2022 Sep 13. Transl Oncol. 2022. PMID: 36113343 Free PMC article.
Cited by
-
Epigenetic Inactivation of RIPK3-Dependent Necroptosis Augments Cisplatin Chemoresistance in Human Osteosarcoma.Int J Mol Sci. 2025 Apr 18;26(8):3863. doi: 10.3390/ijms26083863. Int J Mol Sci. 2025. PMID: 40332549 Free PMC article.
-
PDK1-dependent metabolic reprogramming regulates stemness and tumorigenicity of osteosarcoma stem cells through ATF3.Cell Death Dis. 2025 Jul 29;16(1):574. doi: 10.1038/s41419-025-07903-7. Cell Death Dis. 2025. PMID: 40730548 Free PMC article.
-
Coenzyme Q0 inhibited the NLRP3 inflammasome, metastasis/EMT, and Warburg effect by suppressing hypoxia-induced HIF-1α expression in HNSCC cells.Int J Biol Sci. 2024 May 5;20(8):2790-2813. doi: 10.7150/ijbs.93943. eCollection 2024. Int J Biol Sci. 2024. PMID: 38904007 Free PMC article.
-
The glucose metabolism reprogramming of yak Sertoli cells under hypoxia is regulated by autophagy.BMC Genomics. 2025 Apr 18;26(1):385. doi: 10.1186/s12864-025-11497-x. BMC Genomics. 2025. PMID: 40251498 Free PMC article.
-
Cancer metabolic reprogramming and precision medicine-current perspective.Front Pharmacol. 2024 Oct 17;15:1450441. doi: 10.3389/fphar.2024.1450441. eCollection 2024. Front Pharmacol. 2024. PMID: 39484162 Free PMC article. Review.
References
Publication types
LinkOut - more resources
Full Text Sources

