Genome characterization of influenza A and B viruses in New South Wales, Australia, in 2019: A retrospective study using high-throughput whole genome sequencing
- PMID: 38288510
- PMCID: PMC10824601
- DOI: 10.1111/irv.13252
Genome characterization of influenza A and B viruses in New South Wales, Australia, in 2019: A retrospective study using high-throughput whole genome sequencing
Abstract
Background: During the 2019 severe influenza season, New South Wales (NSW) experienced the highest number of cases in Australia. This study retrospectively investigated the genetic characteristics of influenza viruses circulating in NSW in 2019 and identified genetic markers related to antiviral resistance and potential virulence.
Methods: The complete genomes of influenza A and B viruses were amplified using reverse transcription-polymerase chain reaction (PCR) and sequenced with an Illumina MiSeq platform.
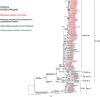
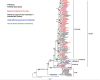
Results: When comparing the sequencing data with the vaccine strains and reference sequences, the phylogenetic analysis revealed that most NSW A/H3N2 viruses (n = 68; 94%) belonged to 3C.2a1b and a minority (n = 4; 6%) belonged to 3C.3a. These viruses all diverged from the vaccine strain A/Switzerland/8060/2017. All A/H1N1pdm09 viruses (n = 20) showed genetic dissimilarity from vaccine strain A/Michigan/45/2015, with subclades 6B.1A.5 and 6B.1A.2 identified. All B/Victoria-lineage viruses (n = 21) aligned with clade V1A.3, presenting triple amino acid deletions at positions 162-164 in the hemagglutinin protein, significantly diverging from the vaccine strain B/Colorado/06/2017. Multiple amino acid substitutions were also found in the internal proteins of influenza viruses, some of which have been previously reported in hospitalized influenza patients in Thailand. Notably, the oseltamivir-resistant marker H275Y was present in one immunocompromised patient infected with A/H1N1pdm09 and the resistance-related mutation I222V was detected in another A/H3N2-infected patient.
Conclusions: Considering antigenic drift and the constant evolution of circulating A and B strains, we believe continuous monitoring of influenza viruses in NSW via the high-throughput sequencing approach provides timely and pivotal information for both public health surveillance and clinical treatment.
Keywords: New South Wales; drug resistance, viral; high-throughput nucleotide sequencing; influenza, human.
© 2024 The Authors. Influenza and Other Respiratory Viruses published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
The resurgence of influenza A/H3N2 virus in Australia after the relaxation of COVID-19 restrictions during the 2022 season.J Med Virol. 2024 Sep;96(9):e29922. doi: 10.1002/jmv.29922. J Med Virol. 2024. PMID: 39295292
-
Genetic characterisation of the influenza viruses circulating in Bulgaria during the 2019-2020 winter season.Virus Genes. 2021 Oct;57(5):401-412. doi: 10.1007/s11262-021-01853-w. Epub 2021 Jun 22. Virus Genes. 2021. PMID: 34156583 Free PMC article.
-
Genetic diversity of influenza A viruses circulating in Bulgaria during the 2018-2019 winter season.J Med Microbiol. 2020 Jul;69(7):986-998. doi: 10.1099/jmm.0.001198. J Med Microbiol. 2020. PMID: 32459617 Free PMC article.
-
Predominance of influenza A(H3N2) viruses during the 2016/2017 season in Bulgaria.J Med Microbiol. 2018 Feb;67(2):228-239. doi: 10.1099/jmm.0.000668. Epub 2018 Jan 3. J Med Microbiol. 2018. PMID: 29297852
-
Predominance of influenza virus A(H3N2) 3C.2a1b and A(H1N1)pdm09 6B.1A5A genetic subclades in the WHO European Region, 2018-2019.Vaccine. 2020 Jul 31;38(35):5707-5717. doi: 10.1016/j.vaccine.2020.06.031. Epub 2020 Jul 3. Vaccine. 2020. PMID: 32624252
Cited by
-
Improved influenza A whole-genome sequencing protocol.Front Cell Infect Microbiol. 2024 Nov 28;14:1497278. doi: 10.3389/fcimb.2024.1497278. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39669272 Free PMC article.
-
Custom barcoded primers for influenza A nanopore sequencing: enhanced performance with reduced preparation time.Front Cell Infect Microbiol. 2025 Apr 15;15:1545032. doi: 10.3389/fcimb.2025.1545032. eCollection 2025. Front Cell Infect Microbiol. 2025. PMID: 40302921 Free PMC article.
-
Antigenic Characterization of H1N1 Influenza Viruses That Circulated During the 2019-2020 Season in Philadelphia, Pennsylvania.Influenza Other Respir Viruses. 2025 Jun;19(6):e70104. doi: 10.1111/irv.70104. Influenza Other Respir Viruses. 2025. PMID: 40452165 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

