The chromosome-level assembly of the wild diploid alfalfa genome provides insights into the full landscape of genomic variations between cultivated and wild alfalfa
- PMID: 38288521
- PMCID: PMC11123407
- DOI: 10.1111/pbi.14300
The chromosome-level assembly of the wild diploid alfalfa genome provides insights into the full landscape of genomic variations between cultivated and wild alfalfa
Abstract
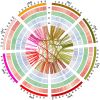
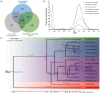
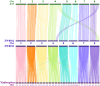
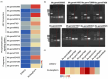
Alfalfa (Medicago sativa L.) is one of the most important forage legumes in the world, including autotetraploid (M. sativa ssp. sativa) and diploid alfalfa (M. sativa ssp. caerulea, progenitor of autotetraploid alfalfa). Here, we reported a high-quality genome of ZW0012 (diploid alfalfa, 769 Mb, contig N50 = 5.5 Mb), which was grouped into the Northern group in population structure analysis, suggesting that our genome assembly filled a major gap among the members of M. sativa complex. During polyploidization, large phenotypic differences occurred between diploids and tetraploids, and the genetic information underlying its massive phenotypic variations remains largely unexplored. Extensive structural variations (SVs) were identified between ZW0012 and XinJiangDaYe (an autotetraploid alfalfa with released genome). We identified 71 ZW0012-specific PAV genes and 1296 XinJiangDaYe-specific PAV genes, mainly involved in defence response, cell growth, and photosynthesis. We have verified the positive roles of MsNCR1 (a XinJiangDaYe-specific PAV gene) in nodulation using an Agrobacterium rhizobia-mediated transgenic method. We also demonstrated that MsSKIP23_1 and MsFBL23_1 (two XinJiangDaYe-specific PAV genes) regulated leaf size by transient overexpression and virus-induced gene silencing analysis. Our study provides a high-quality reference genome of an important diploid alfalfa germplasm and a valuable resource of variation landscape between diploid and autotetraploid, which will facilitate the functional gene discovery and molecular-based breeding for the cultivars in the future.
Keywords: alfalfa; genome assembly; nodule‐specific cysteine‐rich peptides; population structure; structural variants.
© 2024 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



Similar articles
-
A chromosome-scale genome assembly of a diploid alfalfa, the progenitor of autotetraploid alfalfa.Hortic Res. 2020 Dec 1;7(1):194. doi: 10.1038/s41438-020-00417-7. Hortic Res. 2020. PMID: 33328470 Free PMC article.
-
The Chromosome-Level Genome Sequence of the Autotetraploid Alfalfa and Resequencing of Core Germplasms Provide Genomic Resources for Alfalfa Research.Mol Plant. 2020 Sep 7;13(9):1250-1261. doi: 10.1016/j.molp.2020.07.003. Epub 2020 Jul 13. Mol Plant. 2020. PMID: 32673760
-
Inferring population structure and genetic diversity of broad range of wild diploid alfalfa (Medicago sativa L.) accessions using SSR markers.Theor Appl Genet. 2010 Aug;121(3):403-15. doi: 10.1007/s00122-010-1319-4. Epub 2010 Mar 30. Theor Appl Genet. 2010. PMID: 20352180
-
A stepwise guide for pangenome development in crop plants: an alfalfa (Medicago sativa) case study.BMC Genomics. 2024 Oct 31;25(1):1022. doi: 10.1186/s12864-024-10931-w. BMC Genomics. 2024. PMID: 39482604 Free PMC article. Review.
-
Medicago2035: Genomes, functional genomics, and molecular breeding.Mol Plant. 2025 Feb 3;18(2):219-244. doi: 10.1016/j.molp.2024.12.015. Epub 2024 Dec 30. Mol Plant. 2025. PMID: 39741417 Review.
Cited by
-
Functional Genomics: From Soybean to Legume.Int J Mol Sci. 2025 Jun 30;26(13):6323. doi: 10.3390/ijms26136323. Int J Mol Sci. 2025. PMID: 40650105 Free PMC article. Review.
-
QTL detection and genomic prediction for resistance to anthracnose in alfalfa (Medicago sativa).Plant Genome. 2025 Sep;18(3):e70085. doi: 10.1002/tpg2.70085. Plant Genome. 2025. PMID: 40765523 Free PMC article.
-
Advances in basic biology of alfalfa (Medicago sativa L.): a comprehensive overview.Hortic Res. 2025 Mar 10;12(7):uhaf081. doi: 10.1093/hr/uhaf081. eCollection 2025 Jul. Hortic Res. 2025. PMID: 40343348 Free PMC article.
-
Selection and validation of reference genes in alfalfa based on transcriptome sequence data.Sci Rep. 2025 Feb 21;15(1):6324. doi: 10.1038/s41598-025-90664-2. Sci Rep. 2025. PMID: 39984589 Free PMC article.
-
Forage Crop Research in the Modern Age.Adv Sci (Weinh). 2025 Jul;12(27):e2415631. doi: 10.1002/advs.202415631. Epub 2025 Jun 30. Adv Sci (Weinh). 2025. PMID: 40586694 Free PMC article. Review.
References
-
- Allario, T. , Brumos, J. , Colmenero‐Flores, J.M. , Iglesias, D.J. , Pina, J.A. , Navarro, L. , Talon, M. et al. (2013) Tetraploid Rangpur lime rootstock increases drought tolerance via enhanced constitutive root abscisic acid production. Plant Cell Environ. 36, 856–868. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

