Mechanisms of RBM20 Cardiomyopathy: Insights From Model Systems
- PMID: 38288598
- PMCID: PMC10923161
- DOI: 10.1161/CIRCGEN.123.004355
Mechanisms of RBM20 Cardiomyopathy: Insights From Model Systems
Abstract
RBM20 (RNA-binding motif protein 20) is a vertebrate- and muscle-specific RNA-binding protein that belongs to the serine-arginine-rich family of splicing factors. The RBM20 gene was first identified as a dilated cardiomyopathy-linked gene over a decade ago. Early studies in Rbm20 knockout rodents implicated disrupted splicing of RBM20 target genes as a causative mechanism. Clinical studies show that pathogenic variants in RBM20 are linked to aggressive dilated cardiomyopathy with early onset heart failure and high mortality. Subsequent studies employing pathogenic variant knock-in animal models revealed that variants in a specific portion of the arginine-serine-rich domain in RBM20 not only disrupt splicing but also hinder nucleocytoplasmic transport and lead to the formation of RBM20 biomolecular condensates in the sarcoplasm. Conversely, mice harboring a disease-associated variant in the RRM (RNA recognition motif) do not show evidence of adverse remodeling or exhibit sudden death despite disrupted splicing of RBM20 target genes. Thus, whether disrupted splicing, biomolecular condensates, or both contribute to dilated cardiomyopathy is under debate. Beyond this, additional questions remain, such as whether there is sexual dimorphism in the presentation of RBM20 cardiomyopathy. What are the clinical features of RBM20 cardiomyopathy and why do some individuals develop more severe disease than others? In this review, we summarize the reported observations and discuss potential mechanisms of RBM20 cardiomyopathy derived from studies employing in vivo animal models and in vitro human-induced pluripotent stem cell-derived cardiomyocytes. Potential therapeutic strategies to treat RBM20 cardiomyopathy are also discussed.
Keywords: alternative splicing; biomolecular condensates; cardiomyopathy, dilated; heart failure; induced pluripotent stem cell.
Conflict of interest statement
Figures




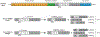
References
-
- Richardson P, McKenna W, Bristow M, Maisch B, Mautner B, O’Connell J, Olsen E, Thiene G, Goodwin J, Gyarfas I, et al. Report of the 1995 World Health Organization/International Society and Federation of Cardiology Task Force on the Definition and Classification of cardiomyopathies. Circulation. 1996;93:841–842. doi: 10.1161/01.cir.93.5.841 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

