Stable population structure in Europe since the Iron Age, despite high mobility
- PMID: 38288729
- PMCID: PMC10827293
- DOI: 10.7554/eLife.79714
Stable population structure in Europe since the Iron Age, despite high mobility
Abstract
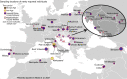
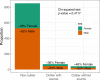
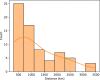
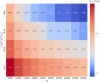
Ancient DNA research in the past decade has revealed that European population structure changed dramatically in the prehistoric period (14,000-3000 years before present, YBP), reflecting the widespread introduction of Neolithic farmer and Bronze Age Steppe ancestries. However, little is known about how population structure changed from the historical period onward (3000 YBP - present). To address this, we collected whole genomes from 204 individuals from Europe and the Mediterranean, many of which are the first historical period genomes from their region (e.g. Armenia and France). We found that most regions show remarkable inter-individual heterogeneity. At least 7% of historical individuals carry ancestry uncommon in the region where they were sampled, some indicating cross-Mediterranean contacts. Despite this high level of mobility, overall population structure across western Eurasia is relatively stable through the historical period up to the present, mirroring geography. We show that, under standard population genetics models with local panmixia, the observed level of dispersal would lead to a collapse of population structure. Persistent population structure thus suggests a lower effective migration rate than indicated by the observed dispersal. We hypothesize that this phenomenon can be explained by extensive transient dispersal arising from drastically improved transportation networks and the Roman Empire's mobilization of people for trade, labor, and military. This work highlights the utility of ancient DNA in elucidating finer scale human population dynamics in recent history.
Keywords: Roman Empire; ancient DNA; evolutionary biology; genetics; genomics; human; population structure.
© 2024, Antonio, Weiß, Gao et al.
Conflict of interest statement
MA, CW, SS, VO, HM, JS, OC, BZ, EP, KÖ, LD, PG, DF, ML, TA, SA, PA, CB, ŽB, AB, LB, PB, EB, DB, FC, MC, DD, ID, KE, MF, AG, GG, FG, SG, ZH, RJ, KV, GJ, IK, KK, HK, AK, SK, MK, VK, MK, DK, FL, ML, SL, TL, TL, WL, AS, VM, YC, NM, IM, NM, BM, LN, MN, AN, CN, JN, AO, JP, LP, DP, KP, SP, MR, MS, AS, AS, TS, MT, IT, DT, JT, DV, TW, DW, MŽ, MF, MČ, MŠ, DB, MN, AC, RP, JP No competing interests declared, ZG Reviewing editor, eLife, MB Affiliated with Palisada Ltd. The author has no financial interests to declare, JB, DL Affiliated with Kaducej Ltd. The author has no financial interests to declare, RM Affiliated with Skupina STIK. The author has no financial interests to declare
Figures














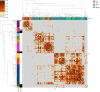

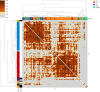
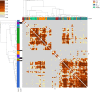







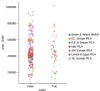




Update of
References
-
- Abulafia D. The Great Sea: A Human History of the Mediterranean. Oxford University Press; 2011. - DOI
-
- Agranat-Tamir L, Waldman S, Martin MAS, Gokhman D, Mishol N, Eshel T, Cheronet O, Rohland N, Mallick S, Adamski N, Lawson AM, Mah M, Michel M, Oppenheimer J, Stewardson K, Candilio F, Keating D, Gamarra B, Tzur S, Novak M, Kalisher R, Bechar S, Eshed V, Kennett DJ, Faerman M, Yahalom-Mack N, Monge JM, Govrin Y, Erel Y, Yakir B, Pinhasi R, Carmi S, Finkelstein I, Carmel L, Reich D. The genomic history of the bronze age southern levant. Cell. 2020;181:1146–1157. doi: 10.1016/j.cell.2020.04.024. - DOI - PMC - PubMed
-
- Allen Ancient DNA Resource Allen ancient DNA resource (AADR): downloadable genotypes of present-day and ancient DNA data. 2021. [May 24, 2021]. https://reich.hms.harvard.edu/allen-ancient-dna-resource-aadr-downloadab...
-
- Allentoft ME, Sikora M, Sjögren KG, Rasmussen S, Rasmussen M, Stenderup J, Damgaard PB, Schroeder H, Ahlström T, Vinner L, Malaspinas AS, Margaryan A, Higham T, Chivall D, Lynnerup N, Harvig L, Baron J, Della Casa P, Dąbrowski P, Duffy PR, Ebel AV, Epimakhov A, Frei K, Furmanek M, Gralak T, Gromov A, Gronkiewicz S, Grupe G, Hajdu T, Jarysz R, Khartanovich V, Khokhlov A, Kiss V, Kolář J, Kriiska A, Lasak I, Longhi C, McGlynn G, Merkevicius A, Merkyte I, Metspalu M, Mkrtchyan R, Moiseyev V, Paja L, Pálfi G, Pokutta D, Pospieszny Ł, Price TD, Saag L, Sablin M, Shishlina N, Smrčka V, Soenov VI, Szeverényi V, Tóth G, Trifanova SV, Varul L, Vicze M, Yepiskoposyan L, Zhitenev V, Orlando L, Sicheritz-Pontén T, Brunak S, Nielsen R, Kristiansen K, Willerslev E. Population genomics of bronze age Eurasia. Nature. 2015;522:167–172. doi: 10.1038/nature14507. - DOI - PubMed
-
- Alpaslan-Roodenberg S, Anthony D, Babiker H, Bánffy E, Booth T, Capone P, Deshpande-Mukherjee A, Eisenmann S, Fehren-Schmitz L, Frachetti M, Fujita R, Frieman CJ, Fu Q, Gibbon V, Haak W, Hajdinjak M, Hofmann KP, Holguin B, Inomata T, Kanzawa-Kiriyama H, Keegan W, Kelso J, Krause J, Kumaresan G, Kusimba C, Kusimba S, Lalueza-Fox C, Llamas B, MacEachern S, Mallick S, Matsumura H, Morales-Arce AY, Matuzeviciute GM, Mushrif-Tripathy V, Nakatsuka N, Nores R, Ogola C, Okumura M, Patterson N, Pinhasi R, Prasad SPR, Prendergast ME, Punzo JL, Reich D, Sawafuji R, Sawchuk E, Schiffels S, Sedig J, Shnaider S, Sirak K, Skoglund P, Slon V, Snow M, Soressi M, Spriggs M, Stockhammer PW, Szécsényi-Nagy A, Thangaraj K, Tiesler V, Tobler R, Wang CC, Warinner C, Yasawardene S, Zahir M. Ethics of DNA research on human remains: five globally applicable guidelines. Nature. 2021;599:41–46. doi: 10.1038/s41586-021-04008-x. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

