Staphylococcus aureus proteases trigger eosinophil-mediated skin inflammation
- PMID: 38289950
- PMCID: PMC10861893
- DOI: 10.1073/pnas.2309243121
Staphylococcus aureus proteases trigger eosinophil-mediated skin inflammation
Abstract
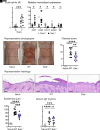
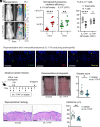
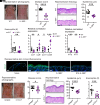
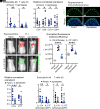
Staphylococcus aureus skin colonization and eosinophil infiltration are associated with many inflammatory skin disorders, including atopic dermatitis, bullous pemphigoid, Netherton's syndrome, and prurigo nodularis. However, whether there is a relationship between S. aureus and eosinophils and how this interaction influences skin inflammation is largely undefined. We show in a preclinical mouse model that S. aureus epicutaneous exposure induced eosinophil-recruiting chemokines and eosinophil infiltration into the skin. Remarkably, we found that eosinophils had a comparable contribution to the skin inflammation as T cells, in a manner dependent on eosinophil-derived IL-17A and IL-17F production. Importantly, IL-36R signaling induced CCL7-mediated eosinophil recruitment to the inflamed skin. Last, S. aureus proteases induced IL-36α expression in keratinocytes, which promoted infiltration of IL-17-producing eosinophils. Collectively, we uncovered a mechanism for S. aureus proteases to trigger eosinophil-mediated skin inflammation, which has implications in the pathogenesis of inflammatory skin diseases.
Keywords: Staphylococcus aureus; eosinophils; inflammatory skin diseases; interleukin-17; interleukin-36.
Conflict of interest statement
Competing interests statement:N.K.A. has received previous grant support from Pfizer and Boehringer Ingelheim and was a paid consultant for Janssen Pharmaceuticals. The remaining authors state no conflict of interest.
Figures



References
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

