A missense SNP in the tumor suppressor SETD2 reduces H3K36me3 and mitotic spindle integrity in Drosophila
- PMID: 38290049
- PMCID: PMC10990431
- DOI: 10.1093/genetics/iyae015
A missense SNP in the tumor suppressor SETD2 reduces H3K36me3 and mitotic spindle integrity in Drosophila
Abstract
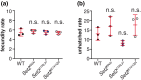
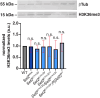
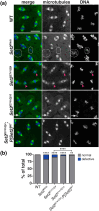
Mutations in SETD2 are among the most prevalent drivers of renal cell carcinoma (RCC). We identified a novel single nucleotide polymorphism (SNP) in SETD2, E902Q, within a subset of RCC patients, which manifests as both an inherited or tumor-associated somatic mutation. To determine if the SNP is biologically functional, we used CRISPR-based genome editing to generate the orthologous mutation within the Drosophila melanogaster Set2 gene. In Drosophila, the homologous amino acid substitution, E741Q, reduces H3K36me3 levels comparable to Set2 knockdown, and this loss is rescued by reintroduction of a wild-type Set2 transgene. We similarly uncovered significant defects in spindle morphogenesis, consistent with the established role of SETD2 in methylating α-Tubulin during mitosis to regulate microtubule dynamics and maintain genome stability. These data indicate the Set2 E741Q SNP affects both histone methylation and spindle integrity. Moreover, this work further suggests the SETD2 E902Q SNP may hold clinical relevance.
Keywords: Drosophila; H3K36me3; SETD2; SNP; models of human disease; renal cell carcinoma.
© The Author(s) 2024. Published by Oxford University Press on behalf of The Genetics Society of America. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Conflict of interest statement
Conflicts of interest The author(s) declare no conflict of interest.
Figures



References
-
- Apweiler R, Attwood TK, Bairoch A, Bateman A, Birney E, Biswas M, Bucher P, Cerutti L, Corpet F, Croning MD, et al. 2000. InterPro–an integrated documentation resource for protein families, domains and functional sites. Bioinformatics. 16(12):1145–1150. doi:10.1093/bioinformatics/16.12.1145. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

