On-Site Fluorescent Detection of Sepsis-Inducing Bacteria using a Graphene-Oxide CRISPR-Cas12a (GO-CRISPR) System
- PMID: 38290431
- PMCID: PMC10867801
- DOI: 10.1021/acs.analchem.3c05459
On-Site Fluorescent Detection of Sepsis-Inducing Bacteria using a Graphene-Oxide CRISPR-Cas12a (GO-CRISPR) System
Abstract
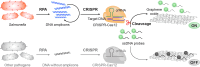
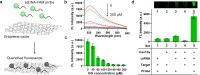
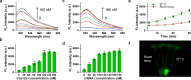
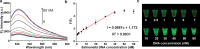
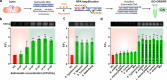
Sepsis is an extremely dangerous medical condition that emanates from the body's response to a pre-existing infection. Early detection of sepsis-inducing bacterial infections can greatly enhance the treatment process and potentially prevent the onset of sepsis. However, current point-of-care (POC) sensors are often complex and costly or lack the ideal sensitivity for effective bacterial detection. Therefore, it is crucial to develop rapid and sensitive biosensors for the on-site detection of sepsis-inducing bacteria. Herein, we developed a graphene oxide CRISPR-Cas12a (GO-CRISPR) biosensor for the detection of sepsis-inducing bacteria in human serum. In this strategy, single-stranded (ssDNA) FAM probes were quenched with single-layer graphene oxide (GO). Target-activated Cas12a trans-cleavage was utilized for the degradation of the ssDNA probes, detaching the short ssDNA probes from GO and recovering the fluorescent signals. Under optimal conditions, we employed our GO-CRISPR system for the detection of Salmonella Typhimurium (S. Typhimurium) with a detection sensitivity of as low as 3 × 103 CFU/mL in human serum, as well as a good detection specificity toward other competing bacteria. In addition, the GO-CRISPR biosensor exhibited excellent sensitivity to the detection of S. Typhimurium in spiked human serum. The GO-CRISPR system offers superior rapidity for the detection of sepsis-inducing bacteria and has the potential to enhance the early detection of bacterial infections in resource-limited settings, expediting the response for patients at risk of sepsis.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

Similar articles
-
Exploring a CRISPR/Cas12a-powered impedimetric biosensor for amplification-free detection of a pathogenic bacterial DNA.Biosens Bioelectron. 2025 Oct 1;285:117607. doi: 10.1016/j.bios.2025.117607. Epub 2025 May 26. Biosens Bioelectron. 2025. PMID: 40419416
-
A ratiometric fluorescent biosensing platform for ultrasensitive detection of Salmonella typhimurium via CRISPR/Cas12a and silver nanoclusters.J Hazard Mater. 2023 Feb 5;443(Pt B):130234. doi: 10.1016/j.jhazmat.2022.130234. Epub 2022 Oct 27. J Hazard Mater. 2023. PMID: 36372024
-
CRISPR-Cas12a Biosensor Array for Ultrasensitive Detection of Unamplified DNA with Single-Nucleotide Polymorphic Discrimination.ACS Sens. 2023 Apr 28;8(4):1489-1499. doi: 10.1021/acssensors.2c02495. Epub 2023 Apr 7. ACS Sens. 2023. PMID: 37027291
-
Application of CRISPR/Cas12a in the rapid detection of pathogens.Clin Chim Acta. 2023 Aug 1;548:117520. doi: 10.1016/j.cca.2023.117520. Epub 2023 Aug 16. Clin Chim Acta. 2023. PMID: 37595863 Review.
-
Sensitive and Portable Signal Readout Strategies Boost Point-of-Care CRISPR/Cas12a Biosensors.ACS Sens. 2023 Nov 24;8(11):3988-4007. doi: 10.1021/acssensors.3c01338. Epub 2023 Oct 23. ACS Sens. 2023. PMID: 37870387 Review.
Cited by
-
Application of pre-amplification-based CRISPR-Cas nanostructured biosensors for bacterial detection.Nanomedicine (Lond). 2025 Apr;20(8):903-915. doi: 10.1080/17435889.2025.2476384. Epub 2025 Mar 7. Nanomedicine (Lond). 2025. PMID: 40052226 Review.
-
CRISPR-Based Biosensors for Medical Diagnosis: Readout from Detector-Dependence Detection Toward Naked Eye Detection.Biosensors (Basel). 2024 Jul 28;14(8):367. doi: 10.3390/bios14080367. Biosensors (Basel). 2024. PMID: 39194596 Free PMC article. Review.
-
Novel Anti-CRISPR-Assisted CRISPR Biosensor for Exclusive Detection of Single-Stranded DNA (ssDNA).ACS Sens. 2024 Mar 22;9(3):1162-1167. doi: 10.1021/acssensors.4c00201. Epub 2024 Mar 5. ACS Sens. 2024. PMID: 38442486 Free PMC article.
-
CIMNE-CRISPR: A novel amplification-free diagnostic for rapid early detection of African Swine Fever Virus.Biosens Bioelectron. 2025 Apr 1;273:117154. doi: 10.1016/j.bios.2025.117154. Epub 2025 Jan 10. Biosens Bioelectron. 2025. PMID: 39826273
-
Optical Bionanosensors for Sepsis Diagnostics.Small. 2025 Feb;21(8):e2409042. doi: 10.1002/smll.202409042. Epub 2025 Jan 2. Small. 2025. PMID: 39745136 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

