ANXA2 (annexin A2) is crucial to ATG7-mediated autophagy, leading to tumor aggressiveness in triple-negative breast cancer cells
- PMID: 38290972
- PMCID: PMC10936647
- DOI: 10.1080/15548627.2024.2305063
ANXA2 (annexin A2) is crucial to ATG7-mediated autophagy, leading to tumor aggressiveness in triple-negative breast cancer cells
Abstract
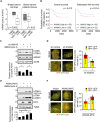
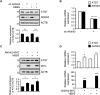
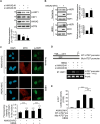
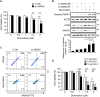
Triple-negative breast cancer (TNBC) is associated with a poor prognosis and metastatic growth. TNBC cells frequently undergo macroautophagy/autophagy, contributing to tumor progression and chemotherapeutic resistance. ANXA2 (annexin A2), a potential therapeutic target for TNBC, has been reported to stimulate autophagy. In this study, we investigated the role of ANXA2 in autophagic processes in TNBC cells. TNBC patients exhibited high levels of ANXA2, which correlated with poor outcomes. ANXA2 increased LC3B-II levels following bafilomycin A1 treatment and enhanced autophagic flux in TNBC cells. Notably, ANXA2 upregulated the phosphorylation of HSF1 (heat shock transcription factor 1), resulting in the transcriptional activation of ATG7 (autophagy related 7). The mechanistic target of rapamycin kinase complex 2 (MTORC2) played an important role in ANXA2-mediated ATG7 transcription by HSF1. MTORC2 did not affect the mRNA level of ANXA2, but it was involved in the protein stability of ANXA2. HSPA (heat shock protein family A (Hsp70)) was a potential interacting protein with ANXA2, which may protect ANXA2 from lysosomal proteolysis. ANXA2 knockdown significantly increased sensitivity to doxorubicin, the first-line chemotherapeutic regimen for TNBC treatment, suggesting that the inhibition of autophagy by ANXA2 knockdown may overcome doxorubicin resistance. In a TNBC xenograft mouse model, we demonstrated that ANXA2 knockdown combined with doxorubicin administration significantly inhibited tumor growth compared to doxorubicin treatment alone, offering a promising avenue to enhance the effectiveness of chemotherapy. In summary, our study elucidated the molecular mechanism by which ANXA2 modulates autophagy, suggesting a potential therapeutic approach for TNBC treatment.Abbreviation: ATG: autophagy related; ChIP: chromatin-immunoprecipitation; HBSS: Hanks' balanced salt solution; HSF1: heat shock transcription factor 1; MTOR: mechanistic target of rapamycin kinase; TNBC: triple-negative breast cancer; TFEB: transcription factor EB; TFE3: transcription factor binding to IGHM enhancer 3.
Keywords: ATG7; Annexin A2; HSF1; MTOR; autophagy; triple-negative breast cancer.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures


References
-
- Mizushima N. Physiological functions of autophagy. Curr Top Microbiol Immunol. 2009;335:71–84. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
