VEXAS syndrome is characterized by inflammasome activation and monocyte dysregulation
- PMID: 38291039
- PMCID: PMC10828464
- DOI: 10.1038/s41467-024-44811-4
VEXAS syndrome is characterized by inflammasome activation and monocyte dysregulation
Abstract
Acquired mutations in the UBA1 gene were recently identified in patients with severe adult-onset auto-inflammatory syndrome called VEXAS (vacuoles, E1 enzyme, X-linked, autoinflammatory, somatic). However, the precise physiological and clinical impact of these mutations remains poorly defined. Here we study a unique prospective cohort of VEXAS patients. We show that monocytes from VEXAS are quantitatively and qualitatively impaired and display features of exhaustion with aberrant expression of chemokine receptors. In peripheral blood from VEXAS patients, we identify an increase in circulating levels of many proinflammatory cytokines, including IL-1β and IL-18 which reflect inflammasome activation and markers of myeloid cells dysregulation. Gene expression analysis of whole blood confirms these findings and also reveals a significant enrichment of TNF-α and NFκB signaling pathways that can mediate cell death and inflammation. This study suggests that the control of the nflammasome activation and inflammatory cell death could be therapeutic targets in VEXAS syndrome.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
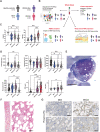
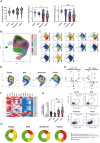

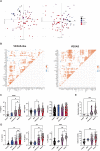
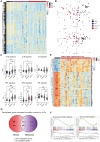
References
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

