Genetic constraints in genes exhibiting splicing plasticity in facultative diapause
- PMID: 38291272
- PMCID: PMC10923799
- DOI: 10.1038/s41437-024-00669-2
Genetic constraints in genes exhibiting splicing plasticity in facultative diapause
Abstract
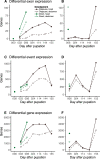
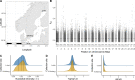
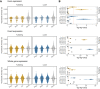
Phenotypic plasticity is produced and maintained by processes regulating the transcriptome. While differential gene expression is among the most important of these processes, relatively little is known about other sources of transcriptional variation. Previous work suggests that alternative splicing plays an extensive and functionally unique role in transcriptional plasticity, though plastically spliced genes may be more constrained than the remainder of expressed genes. In this study, we explore the relationship between expression and splicing plasticity, along with the genetic diversity in those genes, in an ecologically consequential polyphenism: facultative diapause. Using 96 samples spread over two tissues and 10 timepoints, we compare the extent of differential splicing and expression between diapausing and direct developing pupae of the butterfly Pieris napi. Splicing differs strongly between diapausing and direct developing trajectories but alters a smaller and functionally unique set of genes compared to differential expression. We further test the hypothesis that among these expressed loci, plastically spliced genes are likely to experience the strongest purifying selection to maintain seasonally plastic phenotypes. Genes with unique transcriptional changes through diapause consistently had the lowest nucleotide diversity, and this effect was consistently stronger among genes that were differentially spliced compared to those with just differential expression through diapause. Further, the strength of negative selection was higher in the population expressing diapause every generation. Our results suggest that maintenance of the molecular mechanisms involved in diapause progression, including post-transcriptional modifications, are highly conserved and likely to experience genetic constraints, especially in northern populations of P. napi.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Alexa A, Rahnenfuhrer J (2022) topGO: Enrichment Analysis for Gene Ontology. R package version 2.54.0, https://bioconductor.org/packages/topGO
Publication types
MeSH terms
Substances
Grants and funding
- CTS 18:415/Carl Tryggers Stiftelse för Vetenskaplig Forskning (Carl Trygger Foundation)
- 2017-04386/Vetenskapsrådet (Swedish Research Council)
- 2012-3715/Vetenskapsrådet (Swedish Research Council)
- 2010- 5341/Vetenskapsrådet (Swedish Research Council)
- 621-2012-4001/Vetenskapsrådet (Swedish Research Council)
LinkOut - more resources
Full Text Sources

