Cryptosporidium species and subtypes identified in human domestic cases through the national microbiological surveillance programme in Sweden from 2018 to 2022
- PMID: 38291399
- PMCID: PMC10826111
- DOI: 10.1186/s12879-024-09049-x
Cryptosporidium species and subtypes identified in human domestic cases through the national microbiological surveillance programme in Sweden from 2018 to 2022
Abstract
Background: The intestinal protozoan parasite Cryptosporidium is an important cause of diarrheal disease worldwide. A national microbiological surveillance programme was implemented in Sweden in 2018 in order to increase knowledge of the molecular epidemiology of human cryptosporidiosis to better understand transmission patterns and potential zoonotic sources. This article summarises the results of the first five years of the surveillance programme.
Methods: Cryptosporidium-positive faecal and DNA samples from domestically acquired infections were collected from clinical microbiological laboratories in Sweden. Species and subtype determination was performed using 60 kDa glycoprotein and/or small subunit ribosomal RNA gene analysis.
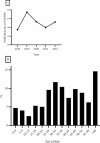
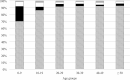
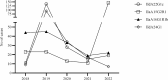
Results: Between 2018 and 2022, 1654 samples were analysed and 11 different species were identified: C. parvum (n = 1412), C. mortiferum (n = 59), C. hominis (n = 56), C. erinacei (n = 11), C. cuniculus (n = 5), C. meleagridis (n = 3), C. equi (n = 2), C. ubiquitum (n = 2), and one each of C. canis, C. ditrichi and C. felis. Subtyping revealed seven subtype families of C. parvum (new subtype families IIy and IIz) and 69 different subtypes (11 new subtypes). The most common C. parvum subtypes were IIdA22G1c, IIdA24G1, IIdA15G2R1 and IIaA16G1R1b. For C. hominis, four different subtype families and nine different subtypes (two new subtypes) were identified. For additional species, two new subtype families (IIIk and VId) and nine new subtypes were identified. All successfully subtyped C. mortiferum cases were subtype XIVaA20G2T1, confirming previous findings in Sweden. Several outbreaks were identified of which the majority were foodborne and a few were due to direct contact with infected animals.
Conclusion: Infection with C. parvum is the leading cause of human cryptosporidiosis acquired in Sweden, where more than 90% of domestic cases are caused by this zoonotic species and only a small proportion of cases are due to infection with other species. The rodent-associated C. mortiferum is considered an emerging zoonotic species in Sweden and the number of domestically acquired human cases has surpassed that of infection with C. hominis. A high diversity of species and subtypes, as well as diversity within the same subtype, was detected. Also, cryptosporidiosis appears to affect adults to a great extent in Sweden.
Keywords: Cryptosporidium; Epidemiology; Molecular typing; Protozoa; Surveillance; Zoonosis.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Tumova L, Jezkova J, Prediger J, Holubova N, Sak B, Konecny R, et al. Cryptosporidium mortiferum n. sp. (Apicomplexa: Cryptosporidiidae), the species causing lethal cryptosporidiosis in eurasian red squirrels (Sciurus vulgaris) Parasit Vectors. 2023;16(1):235. doi: 10.1186/s13071-023-05844-8. - DOI - PMC - PubMed

