This is a preprint.
Alcohol Use Disorder-Associated DNA Methylation in the Nucleus Accumbens and Dorsolateral Prefrontal Cortex
- PMID: 38293028
- PMCID: PMC10827272
- DOI: 10.1101/2024.01.17.23300238
Alcohol Use Disorder-Associated DNA Methylation in the Nucleus Accumbens and Dorsolateral Prefrontal Cortex
Update in
-
Alcohol Use Disorder-Associated DNA Methylation in the Nucleus Accumbens and Dorsolateral Prefrontal Cortex.Biol Psychiatry Glob Open Sci. 2024 Aug 13;4(6):100375. doi: 10.1016/j.bpsgos.2024.100375. eCollection 2024 Nov. Biol Psychiatry Glob Open Sci. 2024. PMID: 39399155 Free PMC article.
Abstract
Background: Alcohol use disorder (AUD) has a profound public health impact. However, understanding of the molecular mechanisms underlying the development and progression of AUD remain limited. Here, we interrogate AUD-associated DNA methylation (DNAm) changes within and across addiction-relevant brain regions: the nucleus accumbens (NAc) and dorsolateral prefrontal cortex (DLPFC).
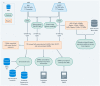
Methods: Illumina HumanMethylation EPIC array data from 119 decedents of European ancestry (61 cases, 58 controls) were analyzed using robust linear regression, with adjustment for technical and biological variables. Associations were characterized using integrative analyses of public gene regulatory data and published genetic and epigenetic studies. We additionally tested for brain region-shared and -specific associations using mixed effects modeling and assessed implications of these results using public gene expression data.
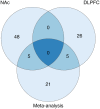
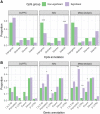
Results: At a false discovery rate ≤ 0.05, we identified 53 CpGs significantly associated with AUD status for NAc and 31 CpGs for DLPFC. In a meta-analysis across the regions, we identified an additional 21 CpGs associated with AUD, for a total of 105 unique AUD-associated CpGs (120 genes). AUD-associated CpGs were enriched in histone marks that tag active promoters and our strongest signals were specific to a single brain region. Of the 120 genes, 23 overlapped with previous genetic associations for substance use behaviors; all others represent novel associations.
Conclusions: Our findings identify AUD-associated methylation signals, the majority of which are specific within NAc or DLPFC. Some signals may constitute predisposing genetic and epigenetic variation, though more work is needed to further disentangle the neurobiological gene regulatory differences associated with AUD.
Keywords: epigenome-wide association study; gene regulation; postmortem human brain; substance use.
Conflict of interest statement
Conflict of Interest The following authors declare no conflict of interest: JDW, MSM, CW, BCQ, SH, RT, AD-S, LZ, SLC, EJCGvdO, TMH, RDM, BTW, EOJ, and DBH. JEK is a member of a drug monitoring committee for an antipsychotic drug trial for Merck. LBJ is listed as an inventor on U.S. Patent 8,080,371, “Markers for Addiction” covering the use of certain SNPs in determining the diagnosis, prognosis, and treatment of addiction.
Figures

References
-
- SAMHSA, Center for Behavioral Health Statistics and Quality. Table 5.6A—Alcohol use disorder in past year: among people aged 12 or older; by age group and demographic characteristics, numbers in thousands, 2021. (2021).
-
- Volkow N. D. & Morales M. The Brain on Drugs: From Reward to Addiction. Cell 162, 712–725 (2015). - PubMed
-
- Grace A. A., Floresco S. B., Goto Y. & Lodge D. J. Regulation of firing of dopaminergic neurons and control of goal-directed behaviors. Trends Neurosci. 30, 220–227 (2007). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
