This is a preprint.
Molecular basis for the transcriptional regulation of an epoxide-based virulence circuit in Pseudomonas aeruginosa
- PMID: 38293063
- PMCID: PMC10827105
- DOI: 10.1101/2024.01.16.572601
Molecular basis for the transcriptional regulation of an epoxide-based virulence circuit in Pseudomonas aeruginosa
Update in
-
Molecular basis for the transcriptional regulation of an epoxide-based virulence circuit in Pseudomonas aeruginosa.Nucleic Acids Res. 2024 Nov 11;52(20):12727-12747. doi: 10.1093/nar/gkae889. Nucleic Acids Res. 2024. PMID: 39413156 Free PMC article.
Abstract
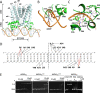
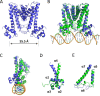
The opportunistic pathogen Pseudomonas aeruginosa infects cystic fibrosis (CF) patient airways and produces a virulence factor Cif that is associated with worse outcomes. Cif is an epoxide hydrolase that reduces cell-surface abundance of the cystic fibrosis transmembrane conductance regulator (CFTR) and sabotages pro-resolving signals. Its expression is regulated by a divergently transcribed TetR family transcriptional repressor. CifR represents the first reported epoxide-sensing bacterial transcriptional regulator, but neither its interaction with cognate operator sequences nor the mechanism of activation has been investigated. Using biochemical and structural approaches, we uncovered the molecular mechanisms controlling this complex virulence operon. We present here the first molecular structures of CifR alone and in complex with operator DNA, resolved in a single crystal lattice. Significant conformational changes between these two structures suggest how CifR regulates the expression of the virulence gene cif. Interactions between the N-terminal extension of CifR with the DNA minor groove of the operator play a significant role in the operator recognition of CifR. We also determined that cysteine residue Cys107 is critical for epoxide sensing and DNA release. These results offer new insights into the stereochemical regulation of an epoxide-based virulence circuit in a critically important clinical pathogen.
Keywords: Epoxide-based virulence circuit; Protein-DNA recognition; Pseudomonas aeruginosa; TetR family transcriptional regulators (TFRs); X-ray crystallography.
Conflict of interest statement
Conflict of Interest: The authors have no conflicts of interest to disclose. CDB owns stock and is an employee of AI Proteins, Inc.
Figures



References
-
- Groenewegen K.H. and Wouters E.F.M. (2003) Bacterial infections in patients requiring admission for an acute exacerbation of COPD; a 1-year prospective study. Respir Med, 97, 770–777. - PubMed
-
- Finch S., Mcdonnell M.J., Abo-leyah H., Aliberti S. and Chalmers J.D. (2015) A Comprehensive Analysis of the Impact of Pseudomonas aeruginosa Colonization on Prognosis in Adult Bronchiectasis. Ann Am Thorac Soc, 12, 1602–1611. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
