This is a preprint.
Binding profiles for 954 Drosophila and C. elegans transcription factors reveal tissue specific regulatory relationships
- PMID: 38293065
- PMCID: PMC10827215
- DOI: 10.1101/2024.01.18.576242
Binding profiles for 954 Drosophila and C. elegans transcription factors reveal tissue specific regulatory relationships
Update in
-
Binding profiles for 961 Drosophila and C. elegans transcription factors reveal tissue-specific regulatory relationships.Genome Res. 2024 Dec 23;34(12):2319-2334. doi: 10.1101/gr.279037.124. Genome Res. 2024. PMID: 39438113 Free PMC article.
Abstract
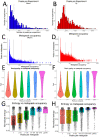
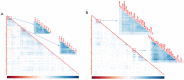
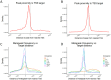
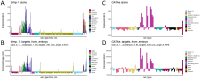
A catalog of transcription factor (TF) binding sites in the genome is critical for deciphering regulatory relationships. Here we present the culmination of the modERN (model organism Encyclopedia of Regulatory Networks) consortium that systematically assayed TF binding events in vivo in two major model organisms, Drosophila melanogaster (fly) and Caenorhabditis elegans (worm). We describe key features of these datasets, comprising 604 TFs identifying 3.6M sites in the fly and 350 TFs identifying 0.9 M sites in the worm. Applying a machine learning model to these data identifies sets of TFs with a prominent role in promoting target gene expression in specific cell types. TF binding data are available through the ENCODE Data Coordinating Center and at https://epic.gs.washington.edu/modERNresource, which provides access to processed and summary data, as well as widgets to probe cell type-specific TF-target relationships. These data are a rich resource that should fuel investigations into TF function during development.
Conflict of interest statement
DECLARATION OF INTERESTS KPW is associated with, and a shareholder in, Tempus Labs and Provaxus, Inc. All other authors do not have external interests.
Figures


References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
