This is a preprint.
Expanding the ligandable proteome by paralog hopping with covalent probes
- PMID: 38293178
- PMCID: PMC10827202
- DOI: 10.1101/2024.01.18.576274
Expanding the ligandable proteome by paralog hopping with covalent probes
Abstract
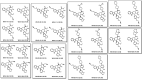
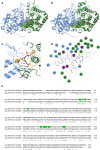
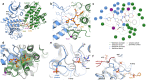
More than half of the ~20,000 protein-encoding human genes have at least one paralog. Chemical proteomics has uncovered many electrophile-sensitive cysteines that are exclusive to a subset of paralogous proteins. Here, we explore whether such covalent compound-cysteine interactions can be used to discover ligandable pockets in paralogs that lack the cysteine. Leveraging the covalent ligandability of C109 in the cyclin CCNE2, we mutated the corresponding residue in paralog CCNE1 to cysteine (N112C) and found through activity-based protein profiling (ABPP) that this mutant reacts stereoselectively and site-specifically with tryptoline acrylamides. We then converted the tryptoline acrylamide-N112C-CCNE1 interaction into a NanoBRET-ABPP assay capable of identifying compounds that reversibly inhibit both N112C- and WT-CCNE1:CDK2 complexes. X-ray crystallography revealed a cryptic allosteric pocket at the CCNE1:CDK2 interface adjacent to N112 that binds the reversible inhibitors. Our findings thus provide a roadmap for leveraging electrophile-cysteine interactions to extend the ligandability of the proteome beyond covalent chemistry.
Figures


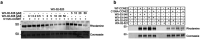










References
-
- Ibn-Salem J. et al. Co-regulation of paralog genes in the three-dimensional chromatin architecture. Nucleic Acids Res 45, 81–91 (2017). https://doi.org: 10.1093/nar/gkw813 - DOI - PMC - PubMed
-
- Man O. et al. From subgenome analysis to protein structure. Curr Opin Struct Biol 13, 353–358 (2003). https://doi.org: 10.1016/s0959-440x(03)00071-x - DOI - PubMed
-
- Conant G. C. et al. Turning a hobby into a job: how duplicated genes find new functions. Nat Rev Genet 9, 938–950 (2008). http://doi.org: 10.1038/nrg2482 - DOI - PubMed
-
- Ryan C. J. et al. Targeting synthetic lethal paralogs in cancer. Trends Cancer 9, 397–409 (2023). https://doi.org: 10.1016/j.trecan.2023.02.002 - DOI - PubMed
-
- Xin Y. et al. Paralog-based synthetic lethality: rationales and applications. Front Oncol 13, 1168143 (2023). https://doi.org: 10.3389/fonc.2023.1168143 - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
