This is a preprint.
DNA Damage Accelerates G-Quadruplex Folding in a Duplex-G-Quadruplex-Duplex Context
- PMID: 38293204
- PMCID: PMC10827223
- DOI: 10.1101/2024.01.20.576387
DNA Damage Accelerates G-Quadruplex Folding in a Duplex-G-Quadruplex-Duplex Context
Update in
-
DNA Damage Accelerates G-Quadruplex Folding in a Duplex-G-Quadruplex-Duplex Context.J Am Chem Soc. 2024 Apr 11;146(16):11364-70. doi: 10.1021/jacs.4c00960. Online ahead of print. J Am Chem Soc. 2024. PMID: 38602473 Free PMC article.
Abstract
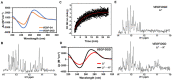
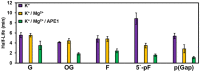
Molecular details for DNA damage impact on the folding of potential G-quadruplex sequences (PQS) to non-canonical DNA structures that are involved in gene regulation are poorly understood. Here, the effects of DNA base damage and strand breaks on PQS folding kinetics were studied in the context of the VEGF promoter sequence embedded between two DNA duplex anchors, referred to as a duplex-G-quadruplex-duplex (DGD) motif. This DGD scaffold imposes constraints on the PQS folding process that more closely mimic those found in genomic DNA. Folding kinetics were monitored by circular dichroism (CD) to find folding half-lives ranging from 2 s to 12 min depending on the DNA damage type and sequence position. The presence of Mg2+ ions and the G-quadruplex (G4)-binding protein APE1 facilitated the folding reactions. A strand break placing all four G runs required for G4 formation on one side of the break accelerated the folding rate by >150-fold compared to the undamaged sequence. Combined 1D 1H-NMR and CD analyses confirmed that isothermal folding of the VEGF-DGD constructs yielded spectral signatures that suggest formation of G4 motifs, and demonstrated a folding dependency with the nature and location of DNA damage. Importantly, the PQS folding half-lives measured are relevant to replication, transcription, and DNA repair time frames.
Figures



Similar articles
-
DNA Damage Accelerates G-Quadruplex Folding in a Duplex-G-Quadruplex-Duplex Context.J Am Chem Soc. 2024 Apr 11;146(16):11364-70. doi: 10.1021/jacs.4c00960. Online ahead of print. J Am Chem Soc. 2024. PMID: 38602473 Free PMC article.
-
Facilitation of a structural transition in the polypurine/polypyrimidine tract within the proximal promoter region of the human VEGF gene by the presence of potassium and G-quadruplex-interactive agents.Nucleic Acids Res. 2005 Oct 20;33(18):6070-80. doi: 10.1093/nar/gki917. Print 2005. Nucleic Acids Res. 2005. PMID: 16239639 Free PMC article.
-
DNA Base Excision Repair Intermediates Influence Duplex-Quadruplex Equilibrium.Molecules. 2023 Jan 18;28(3):970. doi: 10.3390/molecules28030970. Molecules. 2023. PMID: 36770637 Free PMC article.
-
Endogenous oxidized DNA bases and APE1 regulate the formation of G-quadruplex structures in the genome.Proc Natl Acad Sci U S A. 2020 May 26;117(21):11409-11420. doi: 10.1073/pnas.1912355117. Epub 2020 May 13. Proc Natl Acad Sci U S A. 2020. PMID: 32404420 Free PMC article.
-
Structural motifs and intramolecular interactions in non-canonical G-quadruplexes.RSC Chem Biol. 2021 Jan 22;2(2):338-353. doi: 10.1039/d0cb00211a. eCollection 2021 Apr 1. RSC Chem Biol. 2021. PMID: 34458788 Free PMC article. Review.
References
-
- Chambers V. S.; Marsico G.; Boutell J. M.; Di Antonio M.; Smith G. P.; Balasubramanian S., High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat. Biotechnol. 2015, 33, 877–881. - PubMed
-
- Wu J.; McKeague M.; Sturla S. J., Nucleotide-resolution genome-wide mapping of oxidative DNA damage by click-code-seq. J. Am. Chem. Soc. 2018, 140, 9783–9787. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
