This is a preprint.
Multiple Pathways Impact Swarming Motility of Pseudomonas fluorescens Pf0-1
- PMID: 38293239
- PMCID: PMC10827169
- DOI: 10.1101/2024.01.17.576057
Multiple Pathways Impact Swarming Motility of Pseudomonas fluorescens Pf0-1
Update in
-
Multiple pathways impact the swarming motility of Pseudomonas fluorescens Pf0-1.Microbiol Spectr. 2024 Jun 4;12(6):e0016624. doi: 10.1128/spectrum.00166-24. Epub 2024 Apr 30. Microbiol Spectr. 2024. PMID: 38687073 Free PMC article.
Abstract
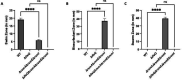
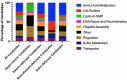
Swarming motility in pseudomonads typically requires both a functional flagellum and production/secretion of a biosurfactant. Published work has shown that the wild-type Pseudomonas fluorescens Pf0-1 is swarming-deficient due to a point mutation in the gacA gene, which until recently, was thought to inactivate rather than attenuate the Gac/Rsm pathway. As a result, little is known about the underlying mechanisms that regulate swarming motility by P. fluorescens Pf0-1. Here, we demonstrate that a ΔrsmA ΔrsmE ΔrsmI mutant, which phenotypically mimics Gac/Rsm pathway overstimulation, is proficient at swarming motility. RsmA and RsmE appear to play a key role in this regulation. Transposon mutagenesis of the ΔrsmA ΔrsmE ΔrsmI mutant identified multiple factors that impact swarming motility, including pathways involved in flagellar synthesis and biosurfactant production/secretion. We find that loss of genes linked to biosurfactant Gacamide A biosynthesis or secretion impact swarming motility, as does loss of the alternative sigma factor FliA, which results in a defect in flagellar function. Collectively, these findings provide evidence that P. fluorescens Pf0-1 can swarm if the Gac/Rsm pathway is activated, highlight the regulatory complexity of swarming motility in this strain, and demonstrate that the cyclic lipopeptide Gacamide A is utilized as a biosurfactant for swarming motility.
Keywords: Pseudomonas fluorescens; biosurfactant; flagellum; regulation; swarming.
Figures




References
-
- Harshey RM. 1994. Bees aren’t the only ones: swarming in gram-negative bacteria. Mol Microbiol 13:389–394. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
