Unusual Base Pair between Two 2-Thiouridines and Its Implication for Nonenzymatic RNA Copying
- PMID: 38293747
- PMCID: PMC10870715
- DOI: 10.1021/jacs.3c11158
Unusual Base Pair between Two 2-Thiouridines and Its Implication for Nonenzymatic RNA Copying
Abstract
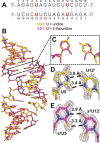
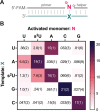
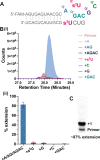
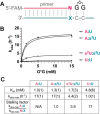
2-Thiouridine (s2U) is a nucleobase modification that confers enhanced efficiency and fidelity both on modern tRNA codon translation and on nonenzymatic and ribozyme-catalyzed RNA copying. We have discovered an unusual base pair between two 2-thiouridines that stabilizes an RNA duplex to a degree that is comparable to that of a native A:U base pair. High-resolution crystal structures indicate similar base-pairing geometry and stacking interactions in duplexes containing s2U:s2U compared to those with U:U pairs. Notably, the C═O···H-N hydrogen bond in the U:U pair is replaced with a C═S···H-N hydrogen bond in the s2U:s2U base pair. The thermodynamic stability of the s2U:s2U base pair suggested that this self-pairing might lead to an increased error frequency during nonenzymatic RNA copying. However, competition experiments show that s2U:s2U base-pairing induces only a low level of misincorporation during nonenzymatic RNA template copying because the correct A:s2U base pair outcompetes the slightly weaker s2U:s2U base pair. In addition, even if an s2U is incorrectly incorporated, the addition of the next base is greatly hindered. This strong stalling effect would further increase the effective fidelity of nonenzymatic RNA copying with s2U. Our findings suggest that s2U may enhance the rate and extent of nonenzymatic copying with only a minimal cost in fidelity.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


References
-
- Grosjean H.; Breton M.; Sirand-Pugnet P.; Tardy F.; Thiaucourt F.; Citti C.; Barré A.; Yoshizawa S.; Fourmy D.; de Crécy-Lagard V.; Blanchard A. Predicting the Minimal Translation Apparatus: Lessons from the Reductive Evolution of Mollicutes. PLoS Genet. 2014, 10 (5), e100436310.1371/journal.pgen.1004363. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

