Focused Screening Identifies Different Sensitivities of Human TET Oxygenases to the Oncometabolite 2-Hydroxyglutarate
- PMID: 38294854
- PMCID: PMC10983004
- DOI: 10.1021/acs.jmedchem.3c01820
Focused Screening Identifies Different Sensitivities of Human TET Oxygenases to the Oncometabolite 2-Hydroxyglutarate
Abstract
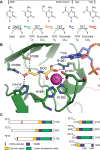
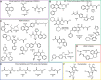
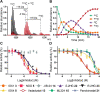
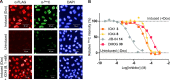
Ten-eleven translocation enzymes (TETs) are Fe(II)/2-oxoglutarate (2OG) oxygenases that catalyze the sequential oxidation of 5-methylcytosine to 5-hydroxymethylcytosine, 5-formylcytosine, and 5-carboxylcytosine in eukaryotic DNA. Despite their roles in epigenetic regulation, there is a lack of reported TET inhibitors. The extent to which 2OG oxygenase inhibitors, including clinically used inhibitors and oncometabolites, modulate DNA modifications via TETs has been unclear. Here, we report studies on human TET1-3 inhibition by a set of 2OG oxygenase-focused inhibitors, employing both enzyme-based and cellular assays. Most inhibitors manifested similar potencies for TET1-3 and caused increases in cellular 5hmC levels. (R)-2-Hydroxyglutarate, an oncometabolite elevated in isocitrate dehydrogenase mutant cancer cells, showed different degrees of inhibition, with TET1 being less potently inhibited than TET3 and TET2, potentially reflecting the proposed role of TET2 mutations in tumorigenesis. The results highlight the tractability of TETs as drug targets and provide starting points for selective inhibitor design.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

References
-
- Tahiliani M.; Koh K. P.; Shen Y.; Pastor W. A.; Bandukwala H.; Brudno Y.; Agarwal S.; Iyer L. M.; Liu D. R.; Aravind L.; Rao A. Conversion of 5-Methylcytosine to 5-Hydroxymethylcytosine in Mammalian DNA by MLL Partner TET1. Science 2009, 324 (5929), 930–935. 10.1126/science.1170116. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Chemical Information
Research Materials

