Rab11 regulates autophagy at dendritic spines in an mTOR- and NMDA-dependent manner
- PMID: 38294869
- PMCID: PMC10916872
- DOI: 10.1091/mbc.E23-02-0060
Rab11 regulates autophagy at dendritic spines in an mTOR- and NMDA-dependent manner
Abstract
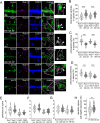
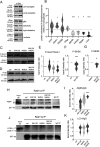
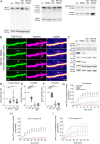
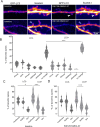
Synaptic plasticity is a process that shapes neuronal connections during neurodevelopment and learning and memory. Autophagy is a mechanism that allows the cell to degrade its unnecessary or dysfunctional components. Autophagosomes appear at dendritic spines in response to plasticity-inducing stimuli. Autophagy defects contribute to altered dendritic spine development, autistic-like behavior in mice, and neurological disease. While several studies have explored the involvement of autophagy in synaptic plasticity, the initial steps of the emergence of autophagosomes at the postsynapse remain unknown. Here, we demonstrate a postsynaptic association of autophagy-related protein 9A (Atg9A), known to be involved in the early stages of autophagosome formation, with Rab11, a small GTPase that regulates endosomal trafficking. Rab11 activity was necessary to maintain Atg9A-positive structures at dendritic spines. Inhibition of mTOR increased Rab11 and Atg9A interaction and increased the emergence of LC3 positive vesicles, an autophagosome membrane-associated protein marker, in dendritic spines when coupled to NMDA receptor stimulation. Dendritic spines with newly formed LC3+ vesicles were more resistant to NMDA-induced morphologic change. Rab11 DN overexpression suppressed appearance of LC3+ vesicles. Collectively, these results suggest that initiation of autophagy in dendritic spines depends on neuronal activity and Rab11a-dependent Atg9A interaction that is regulated by mTOR activity.
Figures


References
-
- Aspernig H, Heimbucher T, Qi W, Gangurde D, Curic S, Yan Y, Donner von Gromoff E, Baumeister R, Thien A (2019). Mitochondrial Perturbations Couple mTORC2 to Autophagy in C. elegans. Cell Reports 29, 1399–1409.e5. - PubMed
-
- Bindels DS, Haarbosch L, van Weeren L, Postma M, Wiese KE, Mastop M, Aumonier S, Gotthard G, Royant A, Hink MA, Gadella TWJ (2017). mScarlet: a bright monomeric red fluorescent protein for cellular imaging. Nat Methods 14, 53–56. - PubMed
-
- Bolte S, Cordelières FP (2006). A guided tour into subcellular colocalization analysis in light microscopy. J Microsc 224, 213–232. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

