Myeloid and lymphoid expression of C9orf72 regulates IL-17A signaling in mice
- PMID: 38295187
- PMCID: PMC11247723
- DOI: 10.1126/scitranslmed.adg7895
Myeloid and lymphoid expression of C9orf72 regulates IL-17A signaling in mice
Abstract
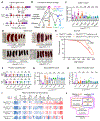
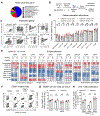
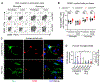
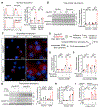
A mutation in C9ORF72 is the most common cause of amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD). Patients with ALS or FTD often develop autoimmunity and inflammation that precedes or coincides with the onset of neurological symptoms, but the underlying mechanisms are poorly understood. Here, we knocked out murine C9orf72 in seven hematopoietic progenitor compartments by conditional mutagenesis and found that myeloid lineage C9orf72 prevents splenomegaly, loss of tolerance, and premature mortality. Furthermore, we demonstrated that C9orf72 plays a role in lymphoid cells to prevent interleukin-17A (IL-17A) production and neutrophilia. Mass cytometry identified early and sustained elevation of the costimulatory molecule CD80 expressed on C9orf72-deficient mouse macrophages, monocytes, and microglia. Enrichment of CD80 was similarly observed in human spinal cord microglia from patients with C9ORF72-mediated ALS compared with non-ALS controls. Single-cell RNA sequencing of murine spinal cord, brain cortex, and spleen demonstrated coordinated induction of gene modules related to antigen processing and presentation and antiviral immunity in C9orf72-deficient endothelial cells, microglia, and macrophages. Mechanistically, C9ORF72 repressed the trafficking of CD80 to the cell surface in response to Toll-like receptor agonists, interferon-γ, and IL-17A. Deletion of Il17a in C9orf72-deficient mice prevented CD80 enrichment in the spinal cord, reduced neutrophilia, and reduced gut T helper type 17 cells. Last, systemic delivery of an IL-17A neutralizing antibody augmented motor performance and suppressed neuroinflammation in C9orf72-deficient mice. Altogether, we show that C9orf72 orchestrates myeloid costimulatory potency and provide support for IL-17A as a therapeutic target for neuroinflammation associated with ALS or FTD.
Conflict of interest statement
Figures



Comment in
-
C9orf72 role in myeloid cells: new perspectives in the investigation of the neuro-immune crosstalk in amyotrophic lateral sclerosis and frontotemporal dementia.Ann Transl Med. 2024 Dec 24;12(6):120. doi: 10.21037/atm-24-86. Epub 2024 Oct 14. Ann Transl Med. 2024. PMID: 39817235 Free PMC article. No abstract available.
References
-
- Majounie E, Renton AE, Mok K, Dopper EGP, Waite A, Rollinson S, Chiò A, Restagno G, Nicolaou N, Simon-Sanchez J, van Swieten JC, Abramzon Y, Johnson JO, Sendtner M, Pamphlett R, Orrell RW, Mead S, Sidle KC, Houlden H, Rohrer JD, Morrison KE, Pall H, Talbot K, Ansorge O, Chromosome 9-ALS/FTD Consortium, French research network on FTLD/FTLD/ALS, ITALSGEN Consortium, Hernandez DG, Arepalli S, Sabatelli M, Mora G, Corbo M, Giannini F, Calvo A, Englund E, Borghero G, Floris GL, Remes AM, Laaksovirta H, McCluskey L, Trojanowski JQ, Van Deerlin VM, Schellenberg GD, Nalls MA, Drory VE, Lu C-S, Yeh T-H, Ishiura H, Takahashi Y, Tsuji S, Le Ber I, Brice A, Drepper C, Williams N, Kirby J, Shaw P, Hardy J, Tienari PJ, Heutink P, Morris HR, Pickering-Brown S, Traynor BJ, Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: a cross-sectional study. Lancet Neurol 11, 323–330 (2012). - PMC - PubMed
-
- Donnelly CJ, Zhang P-W, Pham JT, Haeusler AR, Heusler AR, Mistry NA, Vidensky S, Daley EL, Poth EM, Hoover B, Fines DM, Maragakis N, Tienari PJ, Petrucelli L, Traynor BJ, Wang J, Rigo F, Bennett CF, Blackshaw S, Sattler R, Rothstein JD, RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron 80, 415–428 (2013). - PMC - PubMed
-
- Ash PEA, Bieniek KF, Gendron TF, Caulfield T, Lin W-L, Dejesus-Hernandez M, van Blitterswijk MM, Jansen-West K, Paul JW, Rademakers R, Boylan KB, Dickson DW, Petrucelli L, Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron 77, 639–646 (2013). - PMC - PubMed
-
- Mori K, Weng S-M, Arzberger T, May S, Rentzsch K, Kremmer E, Schmid B, Kretzschmar HA, Cruts M, Van Broeckhoven C, Haass C, Edbauer D, The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science 339, 1335–1338 (2013). - PubMed
-
- Belzil VV, Bauer PO, Prudencio M, Gendron TF, Stetler CT, Yan IK, Pregent L, Daughrity L, Baker MC, Rademakers R, Boylan K, Patel TC, Dickson DW, Petrucelli L, Reduced C9orf72 gene expression in c9FTD/ALS is caused by histone trimethylation, an epigenetic event detectable in blood. Acta Neuropathol 126, 895–905 (2013). - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

