Structural, Biochemical, and Bioinformatic Basis for Identifying Radical SAM Cyclopropyl Synthases
- PMID: 38295270
- PMCID: PMC10878394
- DOI: 10.1021/acschembio.3c00583
Structural, Biochemical, and Bioinformatic Basis for Identifying Radical SAM Cyclopropyl Synthases
Erratum in
-
Correction to "A structural, biochemical, and bioinformatic basis for identifying radical SAM cyclopropyl synthases.".ACS Chem Biol. 2024 May 17;19(5):1206. doi: 10.1021/acschembio.4c00225. Epub 2024 Apr 29. ACS Chem Biol. 2024. PMID: 38683739 Free PMC article. No abstract available.
Abstract
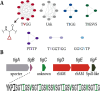
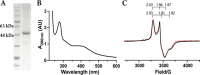
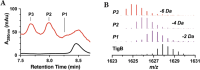
The importance of radical S-adenosyl-l-methionine (RS) enzymes in the maturation of ribosomally synthesized and post-translationally modified peptides (RiPPs) continues to expand, specifically for the RS-SPASM subfamily. We recently discovered an RS-SPASM enzyme that installs a carbon-carbon bond between the geminal methyls of valine residues, resulting in the formation of cyclopropylglycine (CPG). Here, we sought to define the family of cyclopropyl (CP) synthases because of the importance of cyclopropane scaffolds in pharmaceutical development. Using RadicalSAM.org, we bioinformatically expanded the family of CP synthases and assigned unique peptide sequences to each subclade. We identified a unique RiPP biosynthetic pathway that encodes a precursor peptide, TigB, with a repeating TIGSVS motif. Using LCMS and NMR techniques, we show that the RS enzyme associated with the pathway, TigE, catalyzes the formation of a methyl-CPG from the conserved isoleucine residing in the repeating motif of TigB. Furthermore, we obtained a crystal structure of TigE, which reveals an unusual tyrosyl ligation to the auxiliary I [4Fe-4S] cluster, provided by a glycine-tyrosine-tryptophan motif unique to all CP synthases. Further, we show that this unique tyrosyl ligation is absolutely required for TigE activity. Together, our results provide insight into how CP synthases perform this unique reaction.
Conflict of interest statement
The authors declare no competing financial interest.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

