CAG repeat expansions create splicing acceptor sites and produce aberrant repeat-containing RNAs
- PMID: 38295802
- PMCID: PMC10923110
- DOI: 10.1016/j.molcel.2024.01.006
CAG repeat expansions create splicing acceptor sites and produce aberrant repeat-containing RNAs
Abstract
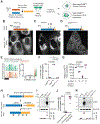
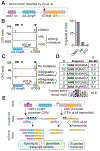
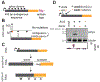
Expansions of CAG trinucleotide repeats cause several rare neurodegenerative diseases. The disease-causing repeats are translated in multiple reading frames and without an identifiable initiation codon. The molecular mechanism of this repeat-associated non-AUG (RAN) translation is not known. We find that expanded CAG repeats create new splice acceptor sites. Splicing of proximal donors to the repeats produces unexpected repeat-containing transcripts. Upon splicing, depending on the sequences surrounding the donor, CAG repeats may become embedded in AUG-initiated open reading frames. Canonical AUG-initiated translation of these aberrant RNAs may account for proteins that have been attributed to RAN translation. Disruption of the relevant splice donors or the in-frame AUG initiation codons is sufficient to abrogate RAN translation. Our findings provide a molecular explanation for the abnormal translation products observed in CAG trinucleotide repeat expansion disorders and add to the repertoire of mechanisms by which repeat expansion mutations disrupt cellular functions.
Keywords: Huntington's disease; RAN translation; RNA splicing; polyglutamine diseases; repeat expansion disorders; repeat-associated non-AUG translation; spinocerebellar ataxia.
Copyright © 2024 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests A.J. is a member of the scientific advisory board of Molecular Cell.
Figures
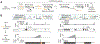


Update of
-
CAG repeat expansions create splicing acceptor sites and produce aberrant repeat-containing RNAs.bioRxiv [Preprint]. 2023 Oct 16:2023.10.16.562581. doi: 10.1101/2023.10.16.562581. bioRxiv. 2023. Update in: Mol Cell. 2024 Feb 15;84(4):702-714.e10. doi: 10.1016/j.molcel.2024.01.006. PMID: 37904984 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

