COQ4 is required for the oxidative decarboxylation of the C1 carbon of coenzyme Q in eukaryotic cells
- PMID: 38295803
- PMCID: PMC7616408
- DOI: 10.1016/j.molcel.2024.01.003
COQ4 is required for the oxidative decarboxylation of the C1 carbon of coenzyme Q in eukaryotic cells
Abstract
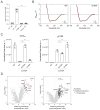
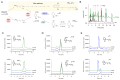
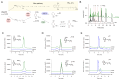
Coenzyme Q (CoQ) is a redox lipid that fulfills critical functions in cellular bioenergetics and homeostasis. CoQ is synthesized by a multi-step pathway that involves several COQ proteins. Two steps of the eukaryotic pathway, the decarboxylation and hydroxylation of position C1, have remained uncharacterized. Here, we provide evidence that these two reactions occur in a single oxidative decarboxylation step catalyzed by COQ4. We demonstrate that COQ4 complements an Escherichia coli strain deficient for C1 decarboxylation and hydroxylation and that COQ4 displays oxidative decarboxylation activity in the non-CoQ producer Corynebacterium glutamicum. Overall, our results substantiate that COQ4 contributes to CoQ biosynthesis, not only via its previously proposed structural role but also via the oxidative decarboxylation of CoQ precursors. These findings fill a major gap in the knowledge of eukaryotic CoQ biosynthesis and shed light on the pathophysiology of human primary CoQ deficiency due to COQ4 mutations.
Keywords: COQ4; Corynebacterium; coenzyme Q; coenzyme Q biosynthesis; coenzyme Q deficiency; mitochondria; oxidative decarboxylation; respiratory chain.
Copyright © 2024 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures

Update of
-
COQ4 is required for the oxidative decarboxylation of the C1 carbon of Coenzyme Q in eukaryotic cells.bioRxiv [Preprint]. 2023 Nov 13:2023.11.13.566839. doi: 10.1101/2023.11.13.566839. bioRxiv. 2023. Update in: Mol Cell. 2024 Mar 7;84(5):981-989.e7. doi: 10.1016/j.molcel.2024.01.003. PMID: 38014142 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

