DENND6A links Arl8b to a Rab34/RILP/dynein complex, regulating lysosomal positioning and autophagy
- PMID: 38296963
- PMCID: PMC10830484
- DOI: 10.1038/s41467-024-44957-1
DENND6A links Arl8b to a Rab34/RILP/dynein complex, regulating lysosomal positioning and autophagy
Abstract
Lysosomes help maintain cellular proteostasis, and defects in lysosomal positioning and function can cause disease, including neurodegenerative disorders. The spatiotemporal distribution of lysosomes is regulated by small GTPases including Rabs, which are activated by guanine nucleotide exchange factors (GEFs). DENN domain proteins are the largest family of Rab GEFs. Using a cell-based assay, we screened DENND6A, a member of the DENN domain protein family against all known Rabs and identified it as a potential GEF for 20 Rabs, including Rab34. Here, we demonstrate that DENND6A activates Rab34, which recruits a RILP/dynein complex to lysosomes, promoting lysosome retrograde transport. Further, we identify DENND6A as an effector of Arl8b, a major regulatory GTPase on lysosomes. We demonstrate that Arl8b recruits DENND6A to peripheral lysosomes to activate Rab34 and initiate retrograde transport, regulating nutrient-dependent lysosomal juxtanuclear repositioning. Loss of DENND6A impairs autophagic flux. Our findings support a model whereby Arl8b/DENND6A/Rab34-dependent lysosomal retrograde trafficking controls autophagy.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
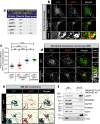
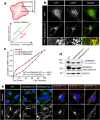
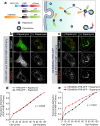
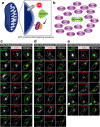
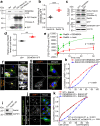
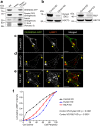
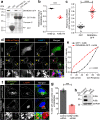
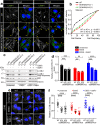

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

