Targeting the m6A RNA methyltransferase METTL3 attenuates the development of kidney fibrosis
- PMID: 38297163
- PMCID: PMC10907702
- DOI: 10.1038/s12276-024-01159-5
Targeting the m6A RNA methyltransferase METTL3 attenuates the development of kidney fibrosis
Abstract
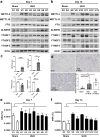
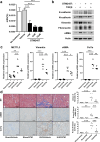
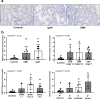
Kidney fibrosis is a major mechanism underlying chronic kidney disease (CKD). N6-methyladenosine (m6A) RNA methylation is associated with organ fibrosis. We investigated m6A profile alterations and the inhibitory effect of RNA methylation in kidney fibrosis in vitro (TGF-β-treated HK-2 cells) and in vivo (unilateral ureteral obstruction [UUO] mouse model). METTL3-mediated signaling was inhibited using siRNA in vitro or the METTL3-specific inhibitor STM2457 in vivo and in vitro. In HK-2 cells, METTL3 protein levels increased in a dose- and time-dependent manner along with an increase in the cellular m6A levels. In the UUO model, METTL3 expression and m6A levels were significantly increased. Transcriptomic and m6A profiling demonstrated that epithelial-to-mesenchymal transition- and inflammation-related pathways were significantly associated with RNA m6A methylation. Genetic and pharmacologic inhibition of METTL3 in HK-2 cells decreased TGF-β-induced fibrotic marker expression. STM2457-induced inhibition of METTL3 attenuated the degree of kidney fibrosis in vivo. Furthermore, METTL3 protein expression was significantly increased in the tissues of CKD patients with diabetic or IgA nephropathy. Therefore, targeting alterations in RNA methylation could be a potential therapeutic strategy for treating kidney fibrosis.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

