BORIS/CTCFL epigenetically reprograms clustered CTCF binding sites into alternative transcriptional start sites
- PMID: 38297316
- PMCID: PMC10832218
- DOI: 10.1186/s13059-024-03175-0
BORIS/CTCFL epigenetically reprograms clustered CTCF binding sites into alternative transcriptional start sites
Abstract
Background: Pervasive usage of alternative promoters leads to the deregulation of gene expression in carcinogenesis and may drive the emergence of new genes in spermatogenesis. However, little is known regarding the mechanisms underpinning the activation of alternative promoters.
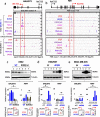
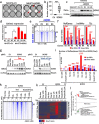
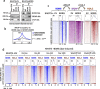
Results: Here we describe how alternative cancer-testis-specific transcription is activated. We show that intergenic and intronic CTCF binding sites, which are transcriptionally inert in normal somatic cells, could be epigenetically reprogrammed into active de novo promoters in germ and cancer cells. BORIS/CTCFL, the testis-specific paralog of the ubiquitously expressed CTCF, triggers the epigenetic reprogramming of CTCF sites into units of active transcription. BORIS binding initiates the recruitment of the chromatin remodeling factor, SRCAP, followed by the replacement of H2A histone with H2A.Z, resulting in a more relaxed chromatin state in the nucleosomes flanking the CTCF binding sites. The relaxation of chromatin around CTCF binding sites facilitates the recruitment of multiple additional transcription factors, thereby activating transcription from a given binding site. We demonstrate that the epigenetically reprogrammed CTCF binding sites can drive the expression of cancer-testis genes, long noncoding RNAs, retro-pseudogenes, and dormant transposable elements.
Conclusions: Thus, BORIS functions as a transcription factor that epigenetically reprograms clustered CTCF binding sites into transcriptional start sites, promoting transcription from alternative promoters in both germ cells and cancer cells.
Keywords: Alternative transcription; CTCF; CTCFL/BORIS; Cancer; Germ cells.
© 2024. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
B.R. is a co-founder and consultant of Arima Genomics Inc. and co-founder of Epigenome Technologies, Inc. The other authors declare that they have no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

