Age-dependent genes in adipose stem and precursor cells affect regulation of fat cell differentiation and link aging to obesity via cellular and genetic interactions
- PMID: 38297378
- PMCID: PMC10829214
- DOI: 10.1186/s13073-024-01291-x
Age-dependent genes in adipose stem and precursor cells affect regulation of fat cell differentiation and link aging to obesity via cellular and genetic interactions
Abstract
Background: Age and obesity are dominant risk factors for several common cardiometabolic disorders, and both are known to impair adipose tissue function. However, the underlying cellular and genetic factors linking aging and obesity on adipose tissue function have remained elusive. Adipose stem and precursor cells (ASPCs) are an understudied, yet crucial adipose cell type due to their deterministic adipocyte differentiation potential, which impacts the capacity to store fat in a metabolically healthy manner.
Methods: We integrated subcutaneous adipose tissue (SAT) bulk (n=435) and large single-nucleus RNA sequencing (n=105) data with the UK Biobank (UKB) (n=391,701) data to study age-obesity interactions originating from ASPCs by performing cell-type decomposition, differential expression testing, cell-cell communication analyses, and construction of polygenic risk scores for body mass index (BMI).
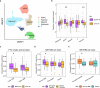
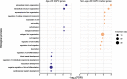
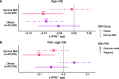
Results: We found that the SAT ASPC proportions significantly decrease with age in an obesity-dependent way consistently in two independent cohorts, both showing that the age dependency of ASPC proportions is abolished by obesity. We further identified 76 genes (72 SAT ASPC marker genes and 4 transcription factors regulating ASPC marker genes) that are differentially expressed by age in SAT and functionally enriched for developmental processes and adipocyte differentiation (i.e., adipogenesis). The 76 age-perturbed ASPC genes include multiple negative regulators of adipogenesis, such as RORA, SMAD3, TWIST2, and ZNF521, form tight clusters of longitudinally co-expressed genes during human adipogenesis, and show age-based differences in cellular interactions between ASPCs and adipose cell types. Finally, our genetic data demonstrate that cis-regional variants of these genes interact with age as predictors of BMI in an obesity-dependent way in the large UKB, while no such gene-age interaction on BMI is observed with non-age-dependent ASPC marker genes, thus independently confirming our cellular ASPC results at the biobank level.
Conclusions: Overall, we discover that obesity prematurely induces a decrease in ASPC proportions and identify 76 developmentally important ASPC genes that implicate altered negative regulation of fat cell differentiation as a mechanism for aging and directly link aging to obesity via significant cellular and genetic interactions.
Keywords: Adipose stem and precursor cells (ASPCs); Aging; Gene-age interactions; Obesity; Polygenic risk score (PRS).
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

