Reduced MUNC18-1 Levels, Synaptic Proteome Changes, and Altered Network Activity in STXBP1-Related Disorder Patient Neurons
- PMID: 38298782
- PMCID: PMC10829628
- DOI: 10.1016/j.bpsgos.2023.05.004
Reduced MUNC18-1 Levels, Synaptic Proteome Changes, and Altered Network Activity in STXBP1-Related Disorder Patient Neurons
Abstract
Background: STXBP1-related disorder (STXBP1-RD) is a neurodevelopmental disorder caused by pathogenic variants in the STXBP1 gene. Its gene product MUNC18-1 organizes synaptic vesicle exocytosis and is essential for synaptic transmission. Patients present with developmental delay, intellectual disability, and/or epileptic seizures, with high clinical heterogeneity. To date, the cellular deficits of neurons of patients with STXBP1-RD are unknown.
Methods: We combined live-cell imaging, electrophysiology, confocal microscopy, and mass spectrometry proteomics to characterize cellular phenotypes of induced pluripotent stem cell-derived neurons from 6 patients with STXBP1-RD, capturing shared features as well as phenotypic diversity among patients.
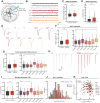
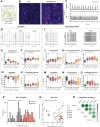
Results: Neurons from all patients showed normal in vitro development, morphology, and synapse formation, but reduced MUNC18-1 RNA and protein levels. In addition, a proteome-wide screen identified dysregulation of proteins related to synapse function and RNA processes. Neuronal networks showed shared as well as patient-specific phenotypes in activity frequency, network irregularity, and synchronicity, especially when networks were challenged by increasing excitability. No shared effects were observed in synapse physiology of single neurons except for a few patient-specific phenotypes. Similarities between functional and proteome phenotypes suggested 2 patient clusters, not explained by gene variant type.
Conclusions: Together, these data show that decreased MUNC18-1 levels, dysregulation of synaptic proteins, and altered network activity are shared cellular phenotypes of STXBP1-RD. The 2 patient clusters suggest distinctive pathobiology among subgroups of patients, providing a plausible explanation for the clinical heterogeneity. This phenotypic spectrum provides a framework for future validation studies and therapy design for STXBP1-RD.
Keywords: MUNC18-1; Neurodevelopmental disorder; SNAREopathy; STXBP1-related disorder; Synaptic transmission; iPSC.
© 2023 The Authors.
Figures




References
-
- Südhof T.C. The molecular machinery of neurotransmitter release (Nobel lecture) Angew Chem Int Ed Engl. 2014;53:12696–12717. - PubMed
-
- Verhage M., Sørensen J.B. SNAREopathies: Diversity in mechanisms and symptoms. Neuron. 2020;107:22–37. - PubMed
-
- Stamberger H., Nikanorova M., Accorsi P., Angriman M., Benkel-Herrenbrueck I., Capovilla G., et al. A neurodevelopmental disorder including epilepsy. Neurology. 2016;86:954–962. - PubMed
-
- Schubert J., Siekierska A., Langlois M., May P., Huneau C., Becker F., et al. Mutations in STX1B, encoding a presynaptic protein, cause fever-associated epilepsy syndromes. Nat Genet. 2014;46:1327–1332. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

